The Role of an Animal-Like Cryptochrome in a Green Alga
 Light is an essential environmental factor for photosynthetic organisms, serving as a source of energy and signal information. To precisely perceive and respond to different wavelengths of the light spectrum, eukaryotic photosynthetic microorganisms and higher plants have developed different classes of light-sensitive receptors, including phototropisns (PHOT), phytochromes and blue-light-absorbing cryptochromes (CRYs). Four CRYs are encoded in the green alga Chlamydomonas reinhardtii, including an animal-like CRY (aCRY) that also absorbs red light. Light conditions influence the sexual life cycle of Chlamydomonas. Illumination, for example, provokes the transition from pregametes to gametes, which achieve full mating ability. The conversion from pregametes to gametes is mainly influenced by blue light, but to some extent also by red light, indicating the participation of blue and/or red light photoreceptors. These considerations led Zou et al. () to investigate whether aCRY is involved in the regulation of the sexual life cycle of C. reinhardtii. They show that aCRY plays an important role in the sexual life cycle of C. reinhardtii. aCRY acts in combination with the C. reinhardtii plant cryptochrome (pCRY) as a negative regulator for mating ability, opposite to the function of phototropin. In contrast, aCRY controls the vegetative germination of the alga in a positive manner, similar to the regulation of this process by PHOT and pCRY.
Light is an essential environmental factor for photosynthetic organisms, serving as a source of energy and signal information. To precisely perceive and respond to different wavelengths of the light spectrum, eukaryotic photosynthetic microorganisms and higher plants have developed different classes of light-sensitive receptors, including phototropisns (PHOT), phytochromes and blue-light-absorbing cryptochromes (CRYs). Four CRYs are encoded in the green alga Chlamydomonas reinhardtii, including an animal-like CRY (aCRY) that also absorbs red light. Light conditions influence the sexual life cycle of Chlamydomonas. Illumination, for example, provokes the transition from pregametes to gametes, which achieve full mating ability. The conversion from pregametes to gametes is mainly influenced by blue light, but to some extent also by red light, indicating the participation of blue and/or red light photoreceptors. These considerations led Zou et al. () to investigate whether aCRY is involved in the regulation of the sexual life cycle of C. reinhardtii. They show that aCRY plays an important role in the sexual life cycle of C. reinhardtii. aCRY acts in combination with the C. reinhardtii plant cryptochrome (pCRY) as a negative regulator for mating ability, opposite to the function of phototropin. In contrast, aCRY controls the vegetative germination of the alga in a positive manner, similar to the regulation of this process by PHOT and pCRY.


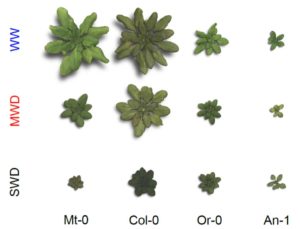 Water deficit (WD) is one of the main environmental stress factors affecting crops and global food security. Acclimation to WD, however, enables plants to maintain growth under unfavorable environmental conditions. To shed light on the molecular mechanisms underlying WD acclimation, Rymaszewski et al. (
Water deficit (WD) is one of the main environmental stress factors affecting crops and global food security. Acclimation to WD, however, enables plants to maintain growth under unfavorable environmental conditions. To shed light on the molecular mechanisms underlying WD acclimation, Rymaszewski et al. (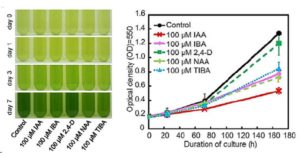 Auxin regulates many aspects of growth and development in land plants, but the origin and evolution of auxin signaling and response mechanisms remain largely unknown. Genome analyses of the moss Physcomitrella patens revealed the presence of the principal gene families involved in auxin homeostasis and signaling in tracheophytes, suggesting that the last common ancestor of land plants had already acquired the core auxin machinery of land plants. However, as we peer further back into phylogenetic time, things get murkier. To address this knowledge deficit, Ohtaka et al. (
Auxin regulates many aspects of growth and development in land plants, but the origin and evolution of auxin signaling and response mechanisms remain largely unknown. Genome analyses of the moss Physcomitrella patens revealed the presence of the principal gene families involved in auxin homeostasis and signaling in tracheophytes, suggesting that the last common ancestor of land plants had already acquired the core auxin machinery of land plants. However, as we peer further back into phylogenetic time, things get murkier. To address this knowledge deficit, Ohtaka et al. (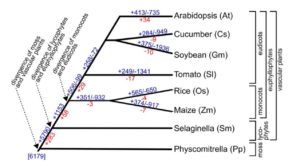 Root hairs are long tubular extensions of root epidermal cells that greatly increase the root surface area and thereby assist in water and nutrient absorption. Root hairs are found in nearly all vascular plants, including angiosperms, gymnosperms, and lycophytes, and they exhibit similar cellular features, suggesting a common evolutionary origin. However, different plant species are known to vary in their root hair distribution patterns and their root hair morphology, implying that genetic differences exist in root hair development programs. Root hairs have been studied intensively in Arabidopsis. In particular, molecular genetic analyses have led to the identification of numerous root hair genes, which provide insight into the mechanisms of Arabidopsis root hair development. Root hair-bearing cells in Arabidopsis are specified by a set of early-acting patterning genes that generate a cell position-dependent distribution of root hair cells and non-hair cells via a complex transcriptional regulatory network. To understand the extent to which this program might operate in other plants, Huang et al. (
Root hairs are long tubular extensions of root epidermal cells that greatly increase the root surface area and thereby assist in water and nutrient absorption. Root hairs are found in nearly all vascular plants, including angiosperms, gymnosperms, and lycophytes, and they exhibit similar cellular features, suggesting a common evolutionary origin. However, different plant species are known to vary in their root hair distribution patterns and their root hair morphology, implying that genetic differences exist in root hair development programs. Root hairs have been studied intensively in Arabidopsis. In particular, molecular genetic analyses have led to the identification of numerous root hair genes, which provide insight into the mechanisms of Arabidopsis root hair development. Root hair-bearing cells in Arabidopsis are specified by a set of early-acting patterning genes that generate a cell position-dependent distribution of root hair cells and non-hair cells via a complex transcriptional regulatory network. To understand the extent to which this program might operate in other plants, Huang et al. (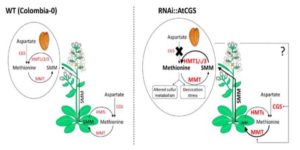 Methionine is a nutritionally essential sulfur-containing amino acid found at low levels in plants and in their seeds. It often limits the nutritional value of crop plants as a source of dietary protein for humans and animals. In plants, methionine plays key roles in protein synthesis and mRNA translation, and regulates indirectly a variety of cellular processes through its main catabolic product S-adenosylmethionine (SAM). SAM serves as the precursor for the synthesis of the plant hormone ethylene, polyamines and biotin, and donates a primary methyl group that is essential for methylation reactions involved in a variety of developmental processes in plant cells. Genetic and biochemical studies suggest that in seeds, methionine can be synthesized de novo as in vegetative tissues via the classical aspartate family pathway by the activity of its main regulatory enzyme, CYSTATHIONINE γ-SYNTHASE (CGS). However, isotope-labeling experiments suggest that methionine can be synthesized in seeds through an alternative pathway by which methionine produced in vegetative tissues is converted to S-methylmethionine (SMM) that is then transported via the phloem into the reproductive tissues where it is converted to methionine. Cohen et al. (
Methionine is a nutritionally essential sulfur-containing amino acid found at low levels in plants and in their seeds. It often limits the nutritional value of crop plants as a source of dietary protein for humans and animals. In plants, methionine plays key roles in protein synthesis and mRNA translation, and regulates indirectly a variety of cellular processes through its main catabolic product S-adenosylmethionine (SAM). SAM serves as the precursor for the synthesis of the plant hormone ethylene, polyamines and biotin, and donates a primary methyl group that is essential for methylation reactions involved in a variety of developmental processes in plant cells. Genetic and biochemical studies suggest that in seeds, methionine can be synthesized de novo as in vegetative tissues via the classical aspartate family pathway by the activity of its main regulatory enzyme, CYSTATHIONINE γ-SYNTHASE (CGS). However, isotope-labeling experiments suggest that methionine can be synthesized in seeds through an alternative pathway by which methionine produced in vegetative tissues is converted to S-methylmethionine (SMM) that is then transported via the phloem into the reproductive tissues where it is converted to methionine. Cohen et al. (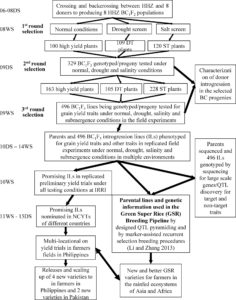 Ali et al. describe a rice breeding strategy to improve abiotic stress tolerance as well as to accelerate the speed to achieving homozygosity. The researchers named this particular technique as “Green Super Rice” (GSR) breeding technology. They use a backcross (BC) breeding approach to fix breeding lines which are tolerant of multiple abiotic stresses and also perform well under normal irrigated conditions to generate large set of improved and tolerant lines to abiotic and biotic stresses (introgression lines). Data from several lines grown in the field are described. (Summary by
Ali et al. describe a rice breeding strategy to improve abiotic stress tolerance as well as to accelerate the speed to achieving homozygosity. The researchers named this particular technique as “Green Super Rice” (GSR) breeding technology. They use a backcross (BC) breeding approach to fix breeding lines which are tolerant of multiple abiotic stresses and also perform well under normal irrigated conditions to generate large set of improved and tolerant lines to abiotic and biotic stresses (introgression lines). Data from several lines grown in the field are described. (Summary by 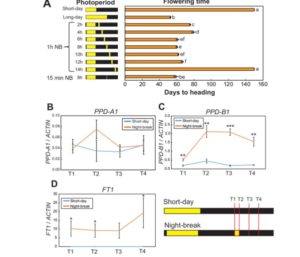 In order to address the knowledge gap in the mechanisms of photoperiodic induction of flowering by phytochrome, Pearce et al. studied flowering behavior in wheat grown under short days, with the interruption of the long nights by short pluses of light (night breaks). Their study showed that night breaks accelerate flowering, and also demonstrated that the response is mediated by photoperiod and phytochrome and requires both PHYB and PHYC. Night breaks affect the expression of PHOTOPERIOD1 (PPD1), which affects the expression of the flowering promoter FT1. (Summary by
In order to address the knowledge gap in the mechanisms of photoperiodic induction of flowering by phytochrome, Pearce et al. studied flowering behavior in wheat grown under short days, with the interruption of the long nights by short pluses of light (night breaks). Their study showed that night breaks accelerate flowering, and also demonstrated that the response is mediated by photoperiod and phytochrome and requires both PHYB and PHYC. Night breaks affect the expression of PHOTOPERIOD1 (PPD1), which affects the expression of the flowering promoter FT1. (Summary by 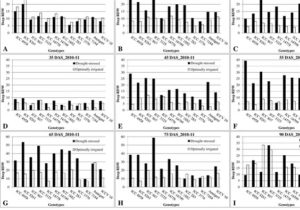 This study by Ramamoorthy et al. showed that survival of plants under drought conditions is not a sufficient goal for breeding. Rather, yield for biomass and food production under water deficit is a better target. Chickpea genotypes having better root growth and higher root density showed better grain filling, and produced good yields in these studies. Some of the genotypes showed proper grain filling with better partition of photosynthates because of higher root density at deeper layers and higher root length at surface. (Summary by
This study by Ramamoorthy et al. showed that survival of plants under drought conditions is not a sufficient goal for breeding. Rather, yield for biomass and food production under water deficit is a better target. Chickpea genotypes having better root growth and higher root density showed better grain filling, and produced good yields in these studies. Some of the genotypes showed proper grain filling with better partition of photosynthates because of higher root density at deeper layers and higher root length at surface. (Summary by 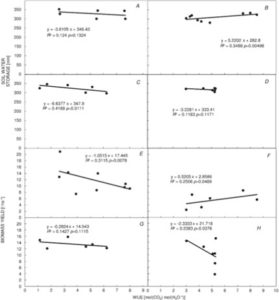 Podlaski et al. conducted experiments with energy crops like miscanthus, prairie cordgrass, willow, etc, and report that water use efficiency (WUE) is no longer a valid trait for selecting energy crops for drought tolerance. They could not find any significant relationship between WUE and biomass yield. However, they found a positive correlation between soil water storage and the biomass yield, and the response was highly differentiated among the crops under various vegetation seasons. (Summary by
Podlaski et al. conducted experiments with energy crops like miscanthus, prairie cordgrass, willow, etc, and report that water use efficiency (WUE) is no longer a valid trait for selecting energy crops for drought tolerance. They could not find any significant relationship between WUE and biomass yield. However, they found a positive correlation between soil water storage and the biomass yield, and the response was highly differentiated among the crops under various vegetation seasons. (Summary by