Eukaryotic pathogens are responsible for devastating plant diseases that threaten food supplies globally – think potato blight caused by the oomycete Phytophora infestans, rice blast caused by the fungus Magnaporthe oryzae, and wheat stem rust caused by the fungus Puccinia graminis f. sp. tritici. These pathogens secrete effector proteins that condition the host cells for successful infection, some by acting in the apoplast and others after entering into the host cells. Many oomycete effectors have an RxLR sequence motif in their N-terminal region that seems to function in host cell targeting, although the mechanisms are a matter of debate (reviewed in Wawra et al., 2012 and Petre and Kamoun, 2014). It is notoriously difficult to study the secretion and targeting of effectors, as these processes occur only at the interface of the pathogen with the host and only during infection. In fact, there are mounting indications that some alternative approaches often used to assess pathogen effector secretion and entry in the host plant could be flawed (see, for example, Petre et al., http://dx.doi.org/10.1101/038232). In a new Breakthrough Report, Wawra & Trusch et al. (2017) provide evidence that the RxLR motif is important for effector secretion from the pathogen, rather than for direct interaction with the host cells.
Plasmodium parasites, which cause malaria, are distantly related to oomycete plant pathogens and similarly have RxLR-like N-terminal sequences that are responsible for targeting to host cells. For many Plasmodium effectors, this so-called PEXEL motif is cleaved in the ER, after which the newly exposed N-terminus is acetylated and the effector is secreted. Another motif, called the TEXEL motif, is important for effector processing in Toxoplasma gondii. The similarity of the RxLR motif to the PEXEL and TEXEL motifs prompted Wawra and coworkers to explore whether RxLR cleavage is involved in effector secretion from P. infestans.
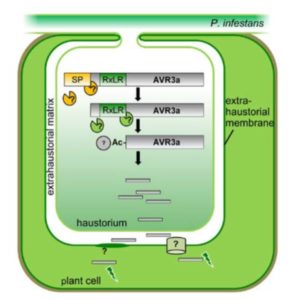 Wawra & Trusch et al. isolated the native (untagged) form of the AVR3a effector secreted into the culture medium by P. infestans. LC-MS/MS analysis did not reveal a peptide representing any sequence more N-terminal than the RxLR motif, but did find a peptide that started immediately downstream of the motif. In addition, the MS data showed likely acetylation. To further characterize the secreted form of the effector, the authors performed reverse-phase chromatography followed by MALDI-TOF analysis. After deglycosylation, the mass of the main product indicated that the AVR3a protein from the medium lacked the first 47 amino acids (i.e., the region up to and including the RxLR motif) with an additional mass for a possible acetylation.
Wawra & Trusch et al. isolated the native (untagged) form of the AVR3a effector secreted into the culture medium by P. infestans. LC-MS/MS analysis did not reveal a peptide representing any sequence more N-terminal than the RxLR motif, but did find a peptide that started immediately downstream of the motif. In addition, the MS data showed likely acetylation. To further characterize the secreted form of the effector, the authors performed reverse-phase chromatography followed by MALDI-TOF analysis. After deglycosylation, the mass of the main product indicated that the AVR3a protein from the medium lacked the first 47 amino acids (i.e., the region up to and including the RxLR motif) with an additional mass for a possible acetylation.
Wawra and coworkers followed up on the possibility of N-terminal acetylation using Edman degradation, which can cleave an N-terminal peptide bond when it is accessible. They observed no cleavage of the AVR3a peptide from the medium, indicating that the N-terminus was not accessible, consistent with it being acetylated. The likely N-acetylation of AVR3a is particularly intriguing as the acetyltransferases that carry out such N-acetylation are found only inside the cell.
Overall, these results are consistent with cleavage of the RxLR motif of AVR3a, followed by acetylation of the new N-terminal amino acid before secretion from P. infestans. It is not clear what protease might be responsible for the cleavage, because none of the 11 P. infestans aspartic proteases homologous to the protease that cleaves the PEXEL motif in Plasmodium could cleave AVR3a in assays using recombinant bacterially expressed proteins. Nevertheless, the potential similarity of this process to the processing and secretion of effectors from other species containing the PEXEL and TEXEL motifs points to its biological relevance and a possible conserved mechanism for effector secretion.
IN BRIEF by Nancy R. Hofmann [email protected]
REFERENCES
Petre, B., and Kamoun, S. (2014). How do filamentous pathogens deliver effector proteins into plant cells? PLoS Biol 12, e1001801.
Wawra, S., Belmonte, R., Lobach, L., Saraiva, M., Willems, A., and van West, P. (2012a). Secretion, delivery and function of oomycete effector proteins. Curr Opin Microbiol 15, 685-691.
Wawra, S., Trusch F., Matena, A., Apostolakis, K., Linne, U., Zhukov, I., Stanek, J., Koźmiński, W., Davidson, I., Secombes, C.J, Bayer, P. and van West, P. (2017). The RxLR Motif of the Host Targeting Effector AVR3a of Phytophthora infestans Is Cleaved Before Secretion. Plant Cell. doi: 10.1105/tpc.16.00552.
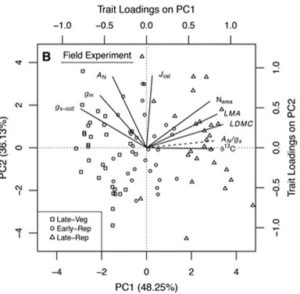 An experimental study by Tomeo and Rosenthal with soybean cultivars demonstrated that there exists genotypic differences in mesophyll conductance (gm), and that the potential exploitation of this trait may increase crop productivity. It was found that there exists a proper coordination mechanism between gm and the physiology of leaf photosynthesis. The gm which alters the CO2 concentration in the leaf photosynthesis apparatus was found to be independent of water loss from the leaf and the exploitation of this trait in selecting genotypes which have higher gm will help in developing improved productive and climate resilient varieties. (Summary by Sridhar Gutam) Plant Physiol.
An experimental study by Tomeo and Rosenthal with soybean cultivars demonstrated that there exists genotypic differences in mesophyll conductance (gm), and that the potential exploitation of this trait may increase crop productivity. It was found that there exists a proper coordination mechanism between gm and the physiology of leaf photosynthesis. The gm which alters the CO2 concentration in the leaf photosynthesis apparatus was found to be independent of water loss from the leaf and the exploitation of this trait in selecting genotypes which have higher gm will help in developing improved productive and climate resilient varieties. (Summary by Sridhar Gutam) Plant Physiol. 

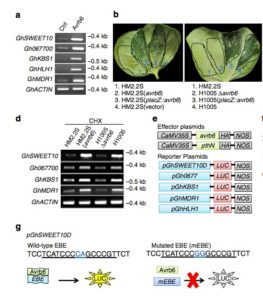 Plants undergo photosynthesis in leaves to produce carbohydrates sucrose and starch. The sucrose is transported to other parts of the plants via sugar transporters called SWEET proteins. In addition, certain plant pathogens activate SWEET genes to invade their host. As shown in this paper, during bacterial blight of cotton (BBC) disease, in cotton GhSWEET10 is induced by Transcription activator-like (TAL) Avrb6 from Xanthomonas citri subsp. malvacearum (Xcm). Understanding this mechanism of infection during BBC would help us develop pathogen resistant varieties of cotton. (Summary by
Plants undergo photosynthesis in leaves to produce carbohydrates sucrose and starch. The sucrose is transported to other parts of the plants via sugar transporters called SWEET proteins. In addition, certain plant pathogens activate SWEET genes to invade their host. As shown in this paper, during bacterial blight of cotton (BBC) disease, in cotton GhSWEET10 is induced by Transcription activator-like (TAL) Avrb6 from Xanthomonas citri subsp. malvacearum (Xcm). Understanding this mechanism of infection during BBC would help us develop pathogen resistant varieties of cotton. (Summary by 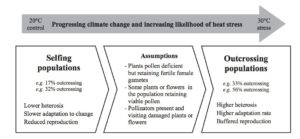 The effects of climate change on agriculture to human health have been well discussed in both scientific and public domains. In plants, changes in climate might affect interactions between the plants and their insect pollinators due to variable availability of pollinators in severe weather conditions, thus affecting reproductive success of plants and crop yield. In this paper, Bishop et al investigate how plant–pollinator interactions are modified by extreme weather by subjecting faba bean (Vicia faba) to altered temperature. Read the paper to see how they quantified pollen movement between plants using a unique phenotypic marker. (Summary by
The effects of climate change on agriculture to human health have been well discussed in both scientific and public domains. In plants, changes in climate might affect interactions between the plants and their insect pollinators due to variable availability of pollinators in severe weather conditions, thus affecting reproductive success of plants and crop yield. In this paper, Bishop et al investigate how plant–pollinator interactions are modified by extreme weather by subjecting faba bean (Vicia faba) to altered temperature. Read the paper to see how they quantified pollen movement between plants using a unique phenotypic marker. (Summary by 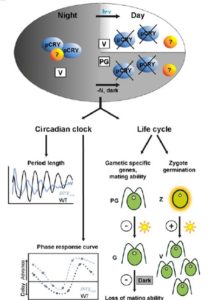 Animals and plants have divergent sets of blue light receptors, called Cryptochromes. However, green alga Chlamydomonas has both animal-like and plant cryptochrome (pCRY). The presence of multiple cryptochrome suggests specific roles in different pathways in respective organisms. In this paper, Müller et al explore the biological functions of pCRY using a mutant with reduced pCRY activity. pcry mutant showed defects in circadian clock with longer period and phase shifts. pcry also show differential expression profile during the life cycle. (Summary by
Animals and plants have divergent sets of blue light receptors, called Cryptochromes. However, green alga Chlamydomonas has both animal-like and plant cryptochrome (pCRY). The presence of multiple cryptochrome suggests specific roles in different pathways in respective organisms. In this paper, Müller et al explore the biological functions of pCRY using a mutant with reduced pCRY activity. pcry mutant showed defects in circadian clock with longer period and phase shifts. pcry also show differential expression profile during the life cycle. (Summary by 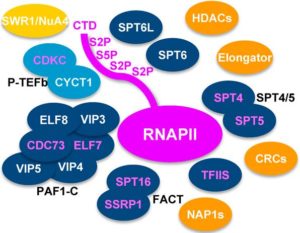 Gene expression is regulated at multiple levels such as genome, transcription, RNA processing and nuclear export, translation, and post-translation. Functional mRNA levels are regulated at transcription stage where RNA polymerase II (RNAPII) controls initiation and elongation of mRNA. In particular, the C-terminal domain of RNAPII defines the elongation phase along with Transcript Elongation Factors (TEFs). Using Reciprocal Tagging approach, Antosz et al identified TEFs in Arabidopsis RNAPII elongation complex. They also studied epistatic interaction between the TEF genes and their effects on plant growth and development. In addition, they co-purified several splicing proteins with TEF-RNAPII complex suggesting interplay between mRNA elongation and processing at the same time. (Summary by
Gene expression is regulated at multiple levels such as genome, transcription, RNA processing and nuclear export, translation, and post-translation. Functional mRNA levels are regulated at transcription stage where RNA polymerase II (RNAPII) controls initiation and elongation of mRNA. In particular, the C-terminal domain of RNAPII defines the elongation phase along with Transcript Elongation Factors (TEFs). Using Reciprocal Tagging approach, Antosz et al identified TEFs in Arabidopsis RNAPII elongation complex. They also studied epistatic interaction between the TEF genes and their effects on plant growth and development. In addition, they co-purified several splicing proteins with TEF-RNAPII complex suggesting interplay between mRNA elongation and processing at the same time. (Summary by 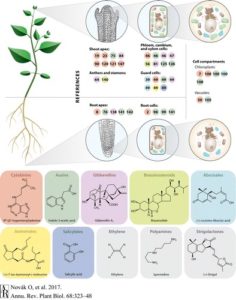 Throughout plant life cycle, from germination till reproduction, every event is regulated by a highly complex network of hormones. Unlike animals where hormones are synthesized in specific glands, each plant cell is able to produce hormones. However, hormones are synthesized in specific organs in plants. Recent advancements in tools have enabled plant scientists to study distribution profiles of phytohormones in plants. Read more about these cool tools in this review by Novák et al. (Summary by
Throughout plant life cycle, from germination till reproduction, every event is regulated by a highly complex network of hormones. Unlike animals where hormones are synthesized in specific glands, each plant cell is able to produce hormones. However, hormones are synthesized in specific organs in plants. Recent advancements in tools have enabled plant scientists to study distribution profiles of phytohormones in plants. Read more about these cool tools in this review by Novák et al. (Summary by  Wawra & Trusch et al. isolated the native (untagged) form of the AVR3a effector secreted into the culture medium by P. infestans. LC-MS/MS analysis did not reveal a peptide representing any sequence more N-terminal than the RxLR motif, but did find a peptide that started immediately downstream of the motif. In addition, the MS data showed likely acetylation. To further characterize the secreted form of the effector, the authors performed reverse-phase chromatography followed by MALDI-TOF analysis. After deglycosylation, the mass of the main product indicated that the AVR3a protein from the medium lacked the first 47 amino acids (i.e., the region up to and including the RxLR motif) with an additional mass for a possible acetylation.
Wawra & Trusch et al. isolated the native (untagged) form of the AVR3a effector secreted into the culture medium by P. infestans. LC-MS/MS analysis did not reveal a peptide representing any sequence more N-terminal than the RxLR motif, but did find a peptide that started immediately downstream of the motif. In addition, the MS data showed likely acetylation. To further characterize the secreted form of the effector, the authors performed reverse-phase chromatography followed by MALDI-TOF analysis. After deglycosylation, the mass of the main product indicated that the AVR3a protein from the medium lacked the first 47 amino acids (i.e., the region up to and including the RxLR motif) with an additional mass for a possible acetylation. Plants have evolved a sophisticated system to defend against pathogen attack that involves recognition of an invading pathogen and activation of the immune system. An increase in calcium concentration in the cell interior is one of the earliest responses of plants exposed to a pathogen attack, and rapid calcium changes are required to the activate the plant immune system. Calcium-dependent protein kinases are calcium sensors and signal transmitters, and are key players in plant immune signaling.
Plants have evolved a sophisticated system to defend against pathogen attack that involves recognition of an invading pathogen and activation of the immune system. An increase in calcium concentration in the cell interior is one of the earliest responses of plants exposed to a pathogen attack, and rapid calcium changes are required to the activate the plant immune system. Calcium-dependent protein kinases are calcium sensors and signal transmitters, and are key players in plant immune signaling.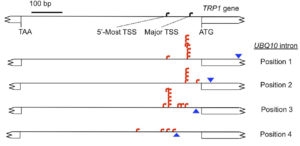 The mechanism remains a mystery, but stimulating introns have a few key features. For example, introns only increase the production of messenger RNA when they are located close to the transcription start site, and they stop working when moved farther away from the start site. Gallegos and Rose set out to define the boundaries of stimulating introns more precisely, to learn more about how introns work to enhance transcription. They found that different positions had different effects on the amount of messenger RNA produced, and also the position at which RNA production started. The authors also tested the importance of the gene promoter region, which normally is thought to exert major control over RNA production. Remarkably, deleting the promoter sequence altogether had no effect on the amount of messenger RNA produced if the gene included a stimulating intron.
The mechanism remains a mystery, but stimulating introns have a few key features. For example, introns only increase the production of messenger RNA when they are located close to the transcription start site, and they stop working when moved farther away from the start site. Gallegos and Rose set out to define the boundaries of stimulating introns more precisely, to learn more about how introns work to enhance transcription. They found that different positions had different effects on the amount of messenger RNA produced, and also the position at which RNA production started. The authors also tested the importance of the gene promoter region, which normally is thought to exert major control over RNA production. Remarkably, deleting the promoter sequence altogether had no effect on the amount of messenger RNA produced if the gene included a stimulating intron.