Mechanisms of Long-Distance mRNA Movement
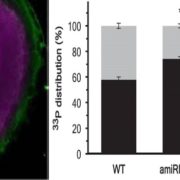
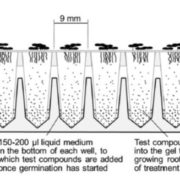
 Phloem has long been recognized as a tissue that transports carbohydrates and amino acids. In recent years, however, it has also been found that this tissue serves as a conduit for signals, e.g., mRNAs, small RNAs, proteins, small peptides, and hormones. Several classical studies have shown that certain mRNAs move from source to sink tissues via the phloem and exert important physiological functions in the distal recipient organs. For example, the development of sink tissues, such as young leaves, tubers, and roots, are partially controlled by the long-distance mobile mRNAs generated in the source tissues. Because a small amount of mobile mRNA is sufficient to generate multiple copies of a protein, such mRNA-based regulatory networks provide efficient ways to incorporate spatial stimuli into development. Two papers in this issue shed light on various facets of long-distance mRNA transport in plants. The first contribution by Xia et al. (10.1104/pp.17.01836) builds on recent heterograft analyses that have shown that large-scale mRNA movement takes place in the phloem. Curiously, however, the number of mobile transcripts reported to be transported in the phloem varies widely, ranging from hundreds to thousands. The authors have developed a heterograft system using Nicotiana benthamiana and tomato (Solanum lycopersicum) to investigate shoot-to-root mRNA movement. Compared to most heterograft systems, the phylogenetic distance between these two species is relatively large, which allowed for more accurate distinction of scion mRNAs from those in the rootstock. The authors report that overall mRNA abundance in the leaf is not a good indicator of transcript mobility to the root, and that increasing the expression levels of non-mobile mRNAs in the companion cells does not promote their mobility. Further underscoring the complexity of long-distance mRNA movements, the authors report that some mRNAs arriving in the roots circulate back to the shoots, and that some mobile mRNAs undergo degradation during their movement.
Phloem has long been recognized as a tissue that transports carbohydrates and amino acids. In recent years, however, it has also been found that this tissue serves as a conduit for signals, e.g., mRNAs, small RNAs, proteins, small peptides, and hormones. Several classical studies have shown that certain mRNAs move from source to sink tissues via the phloem and exert important physiological functions in the distal recipient organs. For example, the development of sink tissues, such as young leaves, tubers, and roots, are partially controlled by the long-distance mobile mRNAs generated in the source tissues. Because a small amount of mobile mRNA is sufficient to generate multiple copies of a protein, such mRNA-based regulatory networks provide efficient ways to incorporate spatial stimuli into development. Two papers in this issue shed light on various facets of long-distance mRNA transport in plants. The first contribution by Xia et al. (10.1104/pp.17.01836) builds on recent heterograft analyses that have shown that large-scale mRNA movement takes place in the phloem. Curiously, however, the number of mobile transcripts reported to be transported in the phloem varies widely, ranging from hundreds to thousands. The authors have developed a heterograft system using Nicotiana benthamiana and tomato (Solanum lycopersicum) to investigate shoot-to-root mRNA movement. Compared to most heterograft systems, the phylogenetic distance between these two species is relatively large, which allowed for more accurate distinction of scion mRNAs from those in the rootstock. The authors report that overall mRNA abundance in the leaf is not a good indicator of transcript mobility to the root, and that increasing the expression levels of non-mobile mRNAs in the companion cells does not promote their mobility. Further underscoring the complexity of long-distance mRNA movements, the authors report that some mRNAs arriving in the roots circulate back to the shoots, and that some mobile mRNAs undergo degradation during their movement.
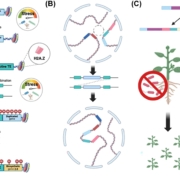
The second contribution in this issue relating to the mRNA mobility in plants is by Luo et al. (10.1104/pp.18.00107), who have examined the question of the mechanism underlying the intracellular sorting of mobile mRNAs. The long-distance trafficking of mobile mRNAs in grafted plants consists of multiple translocation steps, including intracellular trafficking of mobile mRNAs from the nucleus to cytosol, cell-to-cell movement in mesophyll cells and the companion cell/sieve element complex, translocation through phloem, and unloading into and cell-to-cell movement in the destination tissues. Different RNA transport sequences may participate in various steps to determine RNA translocation. Therefore, an approach is needed to effectively investigate the individual translocation steps of mobile mRNA. With this goal in mind, the authors have developed a highly improved fluorescence-based mRNA labeling system, using the bacteriophage coat protein MS2 fused to GFP (MS2-GFP) and an MS2 recognition site in the RNA of interest, to visualize the intracellular trafficking of mobile mRNAs in living plant cells of Nicotiana benthamiana. By means of this RNA imaging system, the authors were able to demonstrate differences in the localization of mobile and nonmobile mRNAs, suggesting that the intracellular targeting of mRNA may be determined by intrinsic mRNA localization signals. Further colocalization studies identified mobile mRNAs targeted to plasmodesmata. These findings suggest that that mobile but not nonmobile mRNAs are selectively targeted to plasmodesmata.