Heterochromatin keeps RNA-directed DNA methylation in place?
Fu et al. demonstrate a requirement for the heterochromatin factors CMT and DDM1 in RNA-directed DNA methylation in maize. Plant Cell https://doi.org/10.1105/tpc.18.00053
By Fang-fang Fu and Jonathan I. Gent
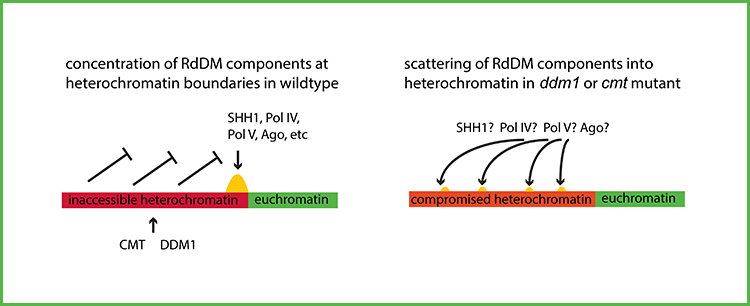
Background: DNA methylation is a chemical modification of DNA consisting of addition of a methyl group to cytosine nucleotides in the DNA molecule. There are three major forms of DNA methylation in vascular plants: (1) replication-coupled CG methylation, (2) chromomethylation, and (3) RNA-directed DNA methylation (RdDM). Chromomethylation is catalyzed by chromomethyltransferases (CMTs), directed by methylation of histone H3 lysine 9, and marks repressed repetitive DNA in heterochromatin. Heterochromatin depends in part on the nucleosome remodeling protein DDM1, which allows access of methyltransferases to nontranscribed, nucleosome-bound DNA. RdDM occurs at heterochromatin boundaries and depends on transcription and a form of RNAi.
Questions: Chromatin modifications associated with heterochromatin promote RdDM, yet heterochromatin itself inhibits RdDM. This apparent contradiction made us wonder what the relationship is between chromomethylatin and RdDM, and whether the relationship differs between maize, whose genome is filled with repetitive DNA, and Arabidopsis, whose genome has little repetitive DNA. In particular we wondered what would happen to RdDM in ddm1 and cmt mutants in maize.
Findings: We found that both ddm1 and cmt mutants survive through endosperm and early embryo development but are nonviable. Whole genome DNA methylation and small RNA sequencing of developing embryo and endosperm revealed dramatic loss of RdDM in mutants. Loss of 24nt small interfering RNA (siRNA) from heterochromatin boundaries was coupled with a gain of 21-, 22-, and 24-nt siRNAs across the repetitive genome, although increased siRNAs were not correlated with increased DNA methylation. Taken together with discoveries from Arabidopsis and rice, we propose a model that when heterochromatin is compromised, RdDM components are scattered from heterochromatin boundaries across the large, repetitive maize genome such that they cannot act in concert for productive RdDM.
Next steps: We predict that any mutants that compromise heterochromatin will also lose RdDM. Currently unavailable but informative mutants in maize would be ones that are defective in histone H3 lysine methylation or in replication-coupled CG methylation.
Fang-Fang Fu, R. Kelly Dawe, Jonathan I. Gent (2018). Loss of RNA-Directed DNA Methylation in Maize Chromomethylase and DDM1-Type Nucleosome Remodeler Mutants. Plant Cell Published July 2018. DOI: https://doi.org/10.1105/tpc.18.00053