What We’re Reading: August 17th
Perspective: The multiplanetary future of plant synthetic biology
 The exploration of space is one of the most inspiring areas of scientific research and a major driver of technological innovation. One of the major factors limiting human expansion trough space is the immensely high cost of resupplying resources from Earth. Mars is the closest neighbor to Earth and although it has the most Earth-like environment, differences in the levels of radiation, soil nutrient content and liquid water availability will limit efficient farming. Using as a framework the forthcoming human mission to Mars, Llorente, Williams and Goold discuss the latest advances in bioengineering that will allow sustainable life beyond Earth, not only by modifying plants’ capacity to seize nutrients in a more efficiently manner, but also by adapting microbes to the extraterrestrial ambient so they can complement and facilitate plant life. (Summary by Aime Jaskalowoski) Gene 10.3390/genes9070348
The exploration of space is one of the most inspiring areas of scientific research and a major driver of technological innovation. One of the major factors limiting human expansion trough space is the immensely high cost of resupplying resources from Earth. Mars is the closest neighbor to Earth and although it has the most Earth-like environment, differences in the levels of radiation, soil nutrient content and liquid water availability will limit efficient farming. Using as a framework the forthcoming human mission to Mars, Llorente, Williams and Goold discuss the latest advances in bioengineering that will allow sustainable life beyond Earth, not only by modifying plants’ capacity to seize nutrients in a more efficiently manner, but also by adapting microbes to the extraterrestrial ambient so they can complement and facilitate plant life. (Summary by Aime Jaskalowoski) Gene 10.3390/genes9070348
A RopGEF regulates asexual reproduction in the liverwort Marchantia polymorpha
 Complex developmental programs regulate tissue and organ formation throughout the green plant lineage, from early diverging non-vascular lineages (bryophytes) to vascular flowering (angiosperm) plants. In the model liverwort Marchantia polymorpha, epidermal patterning gives rise to surface structures such as air pore complexes and gemma cups that house asexually-derived clonal liverwort propagules (gemmae), yet the underlying mechanisms regulating their development remain relatively obscure. In a new bioRxiv pre-print, Hiwatashi et al. (2018) describe the identification of the karappo mutant (meaning “empty”) which develops gemmae cups largely devoid of gemmae. The KARAPPO locus was found to encode a Rop guanine nucleotide exchange factor (GEF) that displays activity on the sole Marchantia Rop GTPase, MpROP. This work identifies KARAPPO/MpRop as a key regulator of the initiation of clonal regeneration in liverworts and provides a framework to assess the conservation of Rop signalling for key asymmetric cell division events in land plants. (Summary by Phil Carella) bioRxiv
Complex developmental programs regulate tissue and organ formation throughout the green plant lineage, from early diverging non-vascular lineages (bryophytes) to vascular flowering (angiosperm) plants. In the model liverwort Marchantia polymorpha, epidermal patterning gives rise to surface structures such as air pore complexes and gemma cups that house asexually-derived clonal liverwort propagules (gemmae), yet the underlying mechanisms regulating their development remain relatively obscure. In a new bioRxiv pre-print, Hiwatashi et al. (2018) describe the identification of the karappo mutant (meaning “empty”) which develops gemmae cups largely devoid of gemmae. The KARAPPO locus was found to encode a Rop guanine nucleotide exchange factor (GEF) that displays activity on the sole Marchantia Rop GTPase, MpROP. This work identifies KARAPPO/MpRop as a key regulator of the initiation of clonal regeneration in liverworts and provides a framework to assess the conservation of Rop signalling for key asymmetric cell division events in land plants. (Summary by Phil Carella) bioRxiv
Nitrogen-fixing corn slime? A Mexican maize landrace supports nitrogen-fixing microbiota in aerial root mucilage
 Plants engage in intimate interactions with symbiotic microbes for the mutually beneficial exchange of nutrients. In a keystone study published in PLoS Biology, Van Deyze et al. (2018) describe the presence of nitrogen-fixing microbiota contained within an extensively carbohydrate rich mucilage found on the aerial roots of a Mexican landrace of maize (Sierra Mixe). Such interactions with nitrogen-fixing microbes are rare in non-leguminous plants (like maize), which further highlights the importance of this unique carbohydrate-for-nitrogen symbiotic transfer in aerial root mucilage. Future efforts to understand the genetic mechanisms regulating this trait in Sierra Mixe maize will aid the eventual biotechnological optimization and transfer to commercial maize varieties that would ideally reduce current dependencies on nitrogen fertilizers. (Summary by Phil Carella) PLOS Bio 10.1371/journal.pbio.2006352
Plants engage in intimate interactions with symbiotic microbes for the mutually beneficial exchange of nutrients. In a keystone study published in PLoS Biology, Van Deyze et al. (2018) describe the presence of nitrogen-fixing microbiota contained within an extensively carbohydrate rich mucilage found on the aerial roots of a Mexican landrace of maize (Sierra Mixe). Such interactions with nitrogen-fixing microbes are rare in non-leguminous plants (like maize), which further highlights the importance of this unique carbohydrate-for-nitrogen symbiotic transfer in aerial root mucilage. Future efforts to understand the genetic mechanisms regulating this trait in Sierra Mixe maize will aid the eventual biotechnological optimization and transfer to commercial maize varieties that would ideally reduce current dependencies on nitrogen fertilizers. (Summary by Phil Carella) PLOS Bio 10.1371/journal.pbio.2006352
Modulating plant growth–metabolism coordination for sustainable agriculture
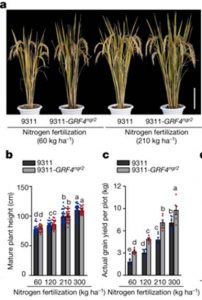 Green revolution varieties of rice and wheat are dwarfed, making them resistant to lodging, and DELLA proteins contribute to this dwarfing. At the same time, green-revolution varieties are not very good at taking up nitrogen, so much of the applied fertilizer is wasted (and polluting). Li et al. have identified and tweaked a control system that enables dwarfed, green-revolution varieties to use nutrients more effectively. A QTL study identified genes that improve nitrogen uptake, one of which encodes the transcription factor GRF4. They found that GRF4 binds to the promoters of several genes involved in N uptake and metabolism, but that its binding is limited by interaction with the DELLA protein SLR1. Alleles of GRF4 that lead to more abundant protein levels lead to enhanced N-use efficiency. They also found that GRF4 regulates genes involved in carbon metabolism (including photosynthesis, sucrose metabolism and sucrose transport) and cell division, also in antagonism with SLR1; as they say, “GRF4–SLR1 antagonism modulates the GA-mediated promotion of cell proliferation, and integrates growth, nitrogen and carbon metabolism regulation.” Green-revolution rice varieties carrying this allele of GRF4 show enhanced grain yields and greater N-use efficiency, without losing the desirable dwarf stature. (Summary by Mary Williams) Nature 10.1038/s41586-018-0415-5
Green revolution varieties of rice and wheat are dwarfed, making them resistant to lodging, and DELLA proteins contribute to this dwarfing. At the same time, green-revolution varieties are not very good at taking up nitrogen, so much of the applied fertilizer is wasted (and polluting). Li et al. have identified and tweaked a control system that enables dwarfed, green-revolution varieties to use nutrients more effectively. A QTL study identified genes that improve nitrogen uptake, one of which encodes the transcription factor GRF4. They found that GRF4 binds to the promoters of several genes involved in N uptake and metabolism, but that its binding is limited by interaction with the DELLA protein SLR1. Alleles of GRF4 that lead to more abundant protein levels lead to enhanced N-use efficiency. They also found that GRF4 regulates genes involved in carbon metabolism (including photosynthesis, sucrose metabolism and sucrose transport) and cell division, also in antagonism with SLR1; as they say, “GRF4–SLR1 antagonism modulates the GA-mediated promotion of cell proliferation, and integrates growth, nitrogen and carbon metabolism regulation.” Green-revolution rice varieties carrying this allele of GRF4 show enhanced grain yields and greater N-use efficiency, without losing the desirable dwarf stature. (Summary by Mary Williams) Nature 10.1038/s41586-018-0415-5
Gating of miRNA movement at defined cell-cell interfaces governs their impact as positional signals
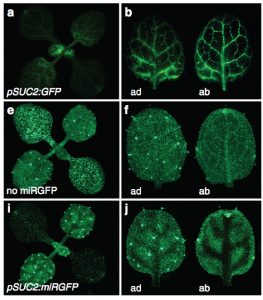
Small RNAs constitute a unique signal transduction mechanism by targeting specific mRNAs and causing a direct down-regulation of target gene expression. Mobile RNAs are known to act as positional cues in developing tissues or to signal stress responses at the systemic level. Although plasmodesmata serve as miRNA gateways in cell-to-cell movement, we know very little about how this movement is regulated. Skopelitis et al. show that the mobility of small RNAs is regulated by a mechanism sensitive to cell polarity, distinct from the one controlling protein movement. The team used a miRGFP sensor system, where tissue-specific expression of miRGFP silences expression of a ubiquitous GFP reporter. The small RNAs were observed to move between cells within a functional domain of the meristem, but not between the domains. Interestingly, the movement of miRNAs in the root meristem was observed between more determined cells, but miRNAs did not move in or out of the quiescent center. The work by Skopelitis et al. provides a framework for future quest in identification of factors controlling polar transport of miRNAs. (Summary by Magdalena Julkowska) Nature Comms. 10.1038/s41467-018-05571-0
Ectopic BASL reveals tissue cell polarity throughout leaf development in Arabidopsis thaliana

CurrBiol Fig.1
Signals determining tissue polarity are fundamental to proper organ development. Asymmetry across the entire organ arises due to to polarity fields. Although there are numerous examples of proteins that express preferential polar localization in the cell, the mechanism that coordinates the polarity field across the whole organ level is still unknown. Mansfield et al. used ectopically expressed BREAKING OF ASYMETRY IN THE STOMATAL LINEAGE (BASL) as a reporter to examine whether the polarity field is dependent on stomatal development. The authors observed asymmetrical BASL localization toward the proximal single lobe. This asymmetric localization was similar in speechless mutant as well as in wild-type plants, suggesting that polarity field is not affected by stomatal development. Cell shape anisotropy, auxin inhibitors or microtubule destabilizing agents did not alter leaf polarity, as reported by BASL. Interestingly, BASL localization was complementary to the PIN1 in developing leaf primodia. In future studies, the newly identified identified proximodistal polarity field might provide an insight into the mechanisms regulating asymmetric organ development. (Summary by Magdalena Julkowska) Curr. Biol. 10.1016/j.cub.2018.06.019
The floral C-lineage genes trigger nectary development in petunia and Arabidopsis
 Nectar production is a crucial feature of plants to attract insects and enhance their expansion across the ecosystems. The molecular basis of nectary development has been studied in detail only in the model plant Arabidopsis thaliana, where the nectaries are positioned on the flower receptacle and near the base of the stamen filaments. In this work, Morel et al. studied the asterid model species Petunia hybrida where the nectaries are tightly associated with the carpels, whose identity depends on the floral homeotic C-function. By analyzing loss-of-function mutants and ectopic expression mutants of two C-function genes, they found that nectary development in petunia and Arabidopsis fully depends on these two genes. Despite the differences in the evolutionary trajectory of the C-linage genes and the different nectary position, the authors conclude that petunia and Arabidopsis employ highly similar molecular mechanisms for nectary development. Finally, Morel et al. show that two petunia genes encoding members of the TOE and AP2 family negatively regulate the size of the nectaries. (Summary by Aime Jaskalowoski) Plant Cell 10.1105/tpc.18.00425
Nectar production is a crucial feature of plants to attract insects and enhance their expansion across the ecosystems. The molecular basis of nectary development has been studied in detail only in the model plant Arabidopsis thaliana, where the nectaries are positioned on the flower receptacle and near the base of the stamen filaments. In this work, Morel et al. studied the asterid model species Petunia hybrida where the nectaries are tightly associated with the carpels, whose identity depends on the floral homeotic C-function. By analyzing loss-of-function mutants and ectopic expression mutants of two C-function genes, they found that nectary development in petunia and Arabidopsis fully depends on these two genes. Despite the differences in the evolutionary trajectory of the C-linage genes and the different nectary position, the authors conclude that petunia and Arabidopsis employ highly similar molecular mechanisms for nectary development. Finally, Morel et al. show that two petunia genes encoding members of the TOE and AP2 family negatively regulate the size of the nectaries. (Summary by Aime Jaskalowoski) Plant Cell 10.1105/tpc.18.00425
Genetic components of water use efficiency in the model grass Setaria
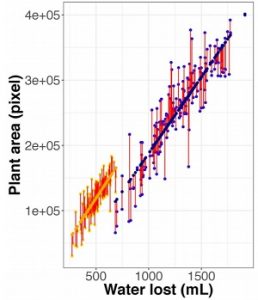 Increasing water use efficiency is important for improving crop yields in diverse environments. This has been difficult due to the complex morphological and biochemical processes involved in water use and plant growth. Feldman and colleagues use a high-throughput phenotyping platform to monitor an interspecific Setaria italica x Setaria viridis recombinant inbred line population under water stress. Plant size and water use were correlated so a linear modeling approach was used to understand deviations from this relationship at the genotypic level. Heritable traits that describe plant size, water use and water use efficiency were found, as well as major loci that underlie two pleiotropic components of water use efficiency. These data can be used to better understand the genetic components of plant size, water use and water use efficiency in grasses. (Summary by Julia Miller) Plant Physiol. 10.1104/pp.18.00146
Increasing water use efficiency is important for improving crop yields in diverse environments. This has been difficult due to the complex morphological and biochemical processes involved in water use and plant growth. Feldman and colleagues use a high-throughput phenotyping platform to monitor an interspecific Setaria italica x Setaria viridis recombinant inbred line population under water stress. Plant size and water use were correlated so a linear modeling approach was used to understand deviations from this relationship at the genotypic level. Heritable traits that describe plant size, water use and water use efficiency were found, as well as major loci that underlie two pleiotropic components of water use efficiency. These data can be used to better understand the genetic components of plant size, water use and water use efficiency in grasses. (Summary by Julia Miller) Plant Physiol. 10.1104/pp.18.00146
Plant acclimation and growth under low temperature conditions mediated by REIL proteins
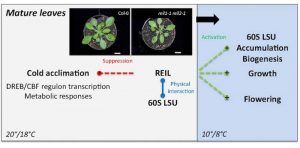 REIL1 and REIL2 proteins in Arabidopsis are homologs of REI1, a yeast ribosome biogenesis factor. REI1 facilitates 60S ribosomal maturation in yeast. Beine-Golovchuk and co-workers have studied the cold acclimation response of the double mutant reil1-1 reil2-1. Previous studies by the same group showed that the double mutant has growth defects below 10°C. In reil1-1 reil1-2, stunted growth was successfully rescued by N-terminal RFP- or GFP-REIL fusion proteins, which allowed the study of REIL1/2 in planta. The FP fusions showed that both REIL1 and REIL2 localized to the cytosol at 20°C and 10°C. One week after the temperature shift to 10°C, Col-0 had and increased amount of cytosolic 60S fraction compared to the plastid ribosomal 50s fraction, while rel1-1 reil2-1 had significantly reduced amounts of 60S fraction. Moreover, western blots of the fractions showed that REIL1 associated with the 60s fraction. Further metabolic and gene expression studies showed that in reil1-1 reil2-1, cold genes and metabolites are induced before the temperature shift, highlighting the importance of REILs to suppress the cold response under non stress temperatures. (Summary by Cecilia Vasquez-Robinet) Plant Physiol. 10.1104/pp.17.01448
REIL1 and REIL2 proteins in Arabidopsis are homologs of REI1, a yeast ribosome biogenesis factor. REI1 facilitates 60S ribosomal maturation in yeast. Beine-Golovchuk and co-workers have studied the cold acclimation response of the double mutant reil1-1 reil2-1. Previous studies by the same group showed that the double mutant has growth defects below 10°C. In reil1-1 reil1-2, stunted growth was successfully rescued by N-terminal RFP- or GFP-REIL fusion proteins, which allowed the study of REIL1/2 in planta. The FP fusions showed that both REIL1 and REIL2 localized to the cytosol at 20°C and 10°C. One week after the temperature shift to 10°C, Col-0 had and increased amount of cytosolic 60S fraction compared to the plastid ribosomal 50s fraction, while rel1-1 reil2-1 had significantly reduced amounts of 60S fraction. Moreover, western blots of the fractions showed that REIL1 associated with the 60s fraction. Further metabolic and gene expression studies showed that in reil1-1 reil2-1, cold genes and metabolites are induced before the temperature shift, highlighting the importance of REILs to suppress the cold response under non stress temperatures. (Summary by Cecilia Vasquez-Robinet) Plant Physiol. 10.1104/pp.17.01448
A conserved role for flavonoids in the protection of plant tissues from UV damage ($)
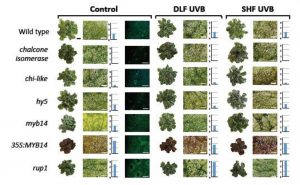 The transition from aquatic to terrestrial environments exposed the earliest land plants to higher doses of damaging ultraviolet (UV-B) radiation. To cope with this stress, land plants evolved complex signalling mechanisms and an inventory of protective ‘sunscreen-like’ flavonoids. To explore whether this paradigm is conserved in extant early divergent land plants, Clayton et al. (2018) investigated the signalling mechanisms induced by UV-B exposure and explored the functional relevance of flavonoid accumulation during UV stress in the model liverwort Marchantia polymorpha. The authors demonstrate that Marchantia employs a UV-B signal transduction cascade similar to that of the model flowering plant Arabidopsis thaliana, through a conserved MpUVR8 UV-B photoreceptor, the transcriptional activator MpHY5 (Elongated Hypocotyl5), and the negative regulator MpRUP1 (Repressor of UV-B Photomorphogenesis1). Moreover, M. polymorpha liverworts exposed to UV-B stress accumulated flavonoids and Marchantia mutants defective in flavonoid production demonstrated increased susceptibility to UV-B damage whereas flavonoid over-accumulation lines (MpMYB14 overexpression) exhibited enhanced tolerance to UV-B. Together these results demonstrate that UV-B signalling modules and flavonoids play a conserved role in UV photoprotection, which suggests that this adaptation likely arose early during the evolution of land plants. (Summary by Phil Carella) Plant J. 10.1111/tpj.14044
The transition from aquatic to terrestrial environments exposed the earliest land plants to higher doses of damaging ultraviolet (UV-B) radiation. To cope with this stress, land plants evolved complex signalling mechanisms and an inventory of protective ‘sunscreen-like’ flavonoids. To explore whether this paradigm is conserved in extant early divergent land plants, Clayton et al. (2018) investigated the signalling mechanisms induced by UV-B exposure and explored the functional relevance of flavonoid accumulation during UV stress in the model liverwort Marchantia polymorpha. The authors demonstrate that Marchantia employs a UV-B signal transduction cascade similar to that of the model flowering plant Arabidopsis thaliana, through a conserved MpUVR8 UV-B photoreceptor, the transcriptional activator MpHY5 (Elongated Hypocotyl5), and the negative regulator MpRUP1 (Repressor of UV-B Photomorphogenesis1). Moreover, M. polymorpha liverworts exposed to UV-B stress accumulated flavonoids and Marchantia mutants defective in flavonoid production demonstrated increased susceptibility to UV-B damage whereas flavonoid over-accumulation lines (MpMYB14 overexpression) exhibited enhanced tolerance to UV-B. Together these results demonstrate that UV-B signalling modules and flavonoids play a conserved role in UV photoprotection, which suggests that this adaptation likely arose early during the evolution of land plants. (Summary by Phil Carella) Plant J. 10.1111/tpj.14044
Suns out, guns out: Plant defense responses are enhanced under long-day photoperiods ($)
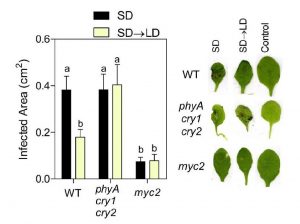 As sessile organisms, plants must constantly sense and respond to a dynamic range of stimuli in their environment, which includes both the duration of light (photoperiod) and the presence of microbial invaders. In a recent article published in Plant Physiology, Cagnola et al. (2018) investigate how plant defense responses are affected in Arabidopsis exposed to increased day length (short to long-day transition). The authors demonstrate that short-day grown plants exposed to a long-day photoperiod activate jasmonic acid-related defense responses that afford enhanced resistance to the necrotrophic fungal pathogen Botrytis cinerea. Genetic analyses further demonstrate that such photoperiod-induced disease resistance is dependent on photoreceptors (phyA/cry1/cry2) and is regulated (negatively) by the master photomorphogenesis regulator COP1. Together these results further corroborate the observation that plant defense responses are tightly linked with environmental conditions in Arabidopsis, which has great implications for crop performance in agricultural fields. (Summary by Phil Carella) Plant Physiol. 10.1104/pp.18.00443
As sessile organisms, plants must constantly sense and respond to a dynamic range of stimuli in their environment, which includes both the duration of light (photoperiod) and the presence of microbial invaders. In a recent article published in Plant Physiology, Cagnola et al. (2018) investigate how plant defense responses are affected in Arabidopsis exposed to increased day length (short to long-day transition). The authors demonstrate that short-day grown plants exposed to a long-day photoperiod activate jasmonic acid-related defense responses that afford enhanced resistance to the necrotrophic fungal pathogen Botrytis cinerea. Genetic analyses further demonstrate that such photoperiod-induced disease resistance is dependent on photoreceptors (phyA/cry1/cry2) and is regulated (negatively) by the master photomorphogenesis regulator COP1. Together these results further corroborate the observation that plant defense responses are tightly linked with environmental conditions in Arabidopsis, which has great implications for crop performance in agricultural fields. (Summary by Phil Carella) Plant Physiol. 10.1104/pp.18.00443
Carbon nanotubes deliver functional genetic material into mature plants without DNA integration
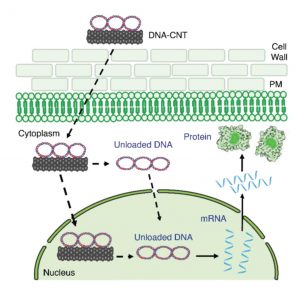 Introducing DNA or RNA into plant cells remains a challenge. et al. describe a new method for transient expression studies through delivery of DNA or RNA via carbon nanotubes (CNTs); the size of the nanoparticles is smaller than the exclusion limit for plant cell walls. The authors show that circular or linear DNA or small RNA can be “grafted” onto CNTs with high efficiency. Using a standard infiltration method (a small nick of the leaf epidermis and injection into leaf tissues using a needle-less syringe), the nanoparticles can readily be introduced into leaf tissues. Using Cy3-tagged CNTs and CNTs conjugated to GFP-encoding DNA, they showed that the DNA-CNT conjugates get into the plant cells and that the DNA is expressed. Expression was high at three days post-infilutration and greatly reduced after ten days, and no integration of DNA into the genome was detected. The authors also demonstrated silencing of constitutively expressed GFP by CNTs grafted to siRNAs targetting GFP. These studies provide a new tool for transient expression or silencing of genes in plants. (Summary by Mary Williams) bioRxiv
Introducing DNA or RNA into plant cells remains a challenge. et al. describe a new method for transient expression studies through delivery of DNA or RNA via carbon nanotubes (CNTs); the size of the nanoparticles is smaller than the exclusion limit for plant cell walls. The authors show that circular or linear DNA or small RNA can be “grafted” onto CNTs with high efficiency. Using a standard infiltration method (a small nick of the leaf epidermis and injection into leaf tissues using a needle-less syringe), the nanoparticles can readily be introduced into leaf tissues. Using Cy3-tagged CNTs and CNTs conjugated to GFP-encoding DNA, they showed that the DNA-CNT conjugates get into the plant cells and that the DNA is expressed. Expression was high at three days post-infilutration and greatly reduced after ten days, and no integration of DNA into the genome was detected. The authors also demonstrated silencing of constitutively expressed GFP by CNTs grafted to siRNAs targetting GFP. These studies provide a new tool for transient expression or silencing of genes in plants. (Summary by Mary Williams) bioRxiv