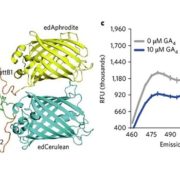
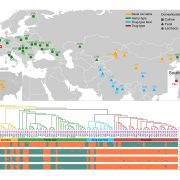
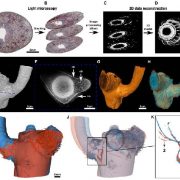
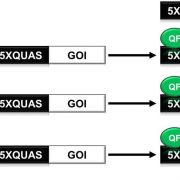
Commentary: Is it ordered correctly? Validating genome assemblies by optical mapping
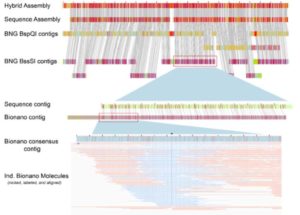 One of the hardest parts of any sequencing project is putting the pieces together. Physical sequence alignments can be cross checked against genetic linkage maps when they are available, but what about for species without genetic linkage data? Udall and Dawe describe the use of optical mapping, using Bionano Genomics or Nabsys technologies, as an additional tool for sequence alignments. These methods use modified restriction enzymes for single-strand nick-based labelling to generate DNA maps. Labeled DNA is passed through nanochannels to generate a map, and the overlapping maps are assembled into an alignment that can be used to validate the physical sequence alignment. In this commentary, the authors describe approaches and limitations to optical mapping for genome assembly. (Summary by Mary Williams) Plant Cell. 10.1105/tpc.17.00514.
One of the hardest parts of any sequencing project is putting the pieces together. Physical sequence alignments can be cross checked against genetic linkage maps when they are available, but what about for species without genetic linkage data? Udall and Dawe describe the use of optical mapping, using Bionano Genomics or Nabsys technologies, as an additional tool for sequence alignments. These methods use modified restriction enzymes for single-strand nick-based labelling to generate DNA maps. Labeled DNA is passed through nanochannels to generate a map, and the overlapping maps are assembled into an alignment that can be used to validate the physical sequence alignment. In this commentary, the authors describe approaches and limitations to optical mapping for genome assembly. (Summary by Mary Williams) Plant Cell. 10.1105/tpc.17.00514.