Clipping Chlamy Genes: Improved Methods for Targeted Gene Editing in Chlamydomonas
A beam of sunlight sends Chlamydomonas reinhardtii scrambling. This tiny, biflagellate alga senses light with its eyespot and adjusts its movements accordingly, depending on photosynthetic needs. In the eyespot, a membranous structure of reddish, carotenoid-filled granules that reflect light and two photoreceptors orchestrate the light guidance of the alga. Two well-characterized photoreceptors, channelrhodopsin 1 (ChR1) and ChR2, act as light-gated ion channels and have been widely used in various animal expression systems. Sixteen other sensory photoreceptor genes have also been identified in Chlamydomonas. Efficiently assessing the roles of sensory photoreceptors in planta requires targeted gene editing, a difficult task in this model system. Homologous recombination between endogenous DNA and homologous DNA templates, which is used to knock out, repair, and modify a gene of interest, rarely occurs in Chlamydomonas because double-strand breaks (DSBs) are usually repaired via error prone non-homologous end joining rather than high-fidelity homology-directed repair (HDR) (Zorin et al., 2005).
The use of sequence-guided nucleases has revolutionized genome editing. Zinc-finger nucleases (ZFNs) contain a DNA cleavage domain fused to a modular array of zinc finger DNA binding domains designed to precisely target the gene of interest. However, the use of ZFNs for gene editing to study photoreceptor (and other) genes in Chlamydomonas has met with limited success. Surprisingly, CRISPR/Cas9 (clustered regularly interspaced short palindromic repeat/associated protein 9), a simpler and cheaper approach for targeted gene editing that has become wildly popular (reviewed in Sander and Joung, 2014), is also inefficient in Chlamydomonas and does not allow for routine disruption of non-selectable genes or genes whose knock-out does not cause a readily identifiable mutant phenotype. The CRISPR system consists of a Cas9 nuclease bound to a single guide RNA (sgRNA) comprising the target sequence (protospacer) joined (by a small linker sequence) to trans-activating CRISPR RNA (which forms a secondary scaffold structure recognized by Cas9). The protospacer portion of the sgRNA directs Cas9 to cleave complementary target-DNA sequences near specific motifs. Designing a ribonucleoprotein (RNP) complex (Cas9 plus sgRNA) for Chlamydomonas is fairly easy, but delivering it (and ZFNs) into this alga is another matter.
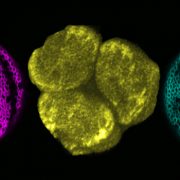
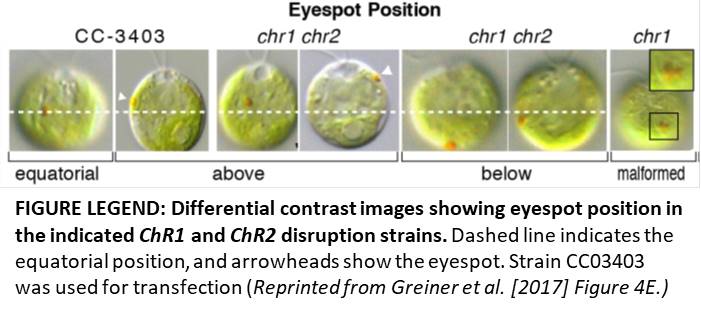
Fortunately, Greiner et al. (2017) greatly improved both processes, providing robust techniques for analyzing the functions of photoreceptor (and other non-selectable) genes in Chlamydomonas and related green algae. The authors created cell wall-deficient Chlamydomonas strains (CC-3403) harboring a mutated antibiotic resistance repair system in their genomes. Marker functionality can only be restored after DSBs are introduced by a correctly functioning ZFN. This elegant strategy allowed them to optimize various experimental parameters, such as heat shock treatment and the use of electroporation rather than glass bead transformation. Using this technique, they successfully targeted ChR1 and ChR2. The eyespots in the resulting chr1 chr2 mutants were often small, mislocalized, and misshapen (see figure), shedding light on the roles of ChR1 and ChR2 in eyespot formation.
The authors then optimized the CRISPR-Cas9 system using a plasmid-based, genetically encoded Cas9, which they used to disrupt four different photoreceptor genes at a high rate. Even better, using an improved method for direct transformation of RNP complexes into Chlamydomonas, eight photoreceptor genes were disrupted in four strains of the alga. In Chlamydomonas, HDR occurs roughly 10-times less often after DNA cleavage with Cas9 versus ZFNs, making ZFNs more suitable for predictably altering genes of interest, although the newly developed techniques are all quite powerful. Now go forth and clip some Chlamy genes.
REFERENCES
Greiner, A., Kelterborn, S., Evers, H., Kreimer, G., Sizova, I., and Hegemann, P. (2017). Targeting of photoreceptor genes via zinc-finger nucleases and CRISPR/Cas9 in Chlamydomonas reinhardtii. Plant Cell 29: doi:10.1015/tpc17.00659.
Sander, J.D. and Joung, J.K. (2014). CriSPr-Cas systems for editing, regulating and targeting genomes. Nature Biotechnology 32: 347–355.
Zorin, B., Hegemann, P., Sizova, I. (2005). Nuclear-gene targeting by using single-stranded DNA avoids illegitimate DNA integration in Chlamydomonas reinhardtii. Eukaryotic Cell 4: 1264-1272. doi:10.1128/EC.4.7.1264-1272.2005.