Making Connections: MAC Function in Splicing and MicroRNA Biogenesis
Fans of the television show The Magic Schoolbus might remember that the teacher/heroine, Ms. Frizzle used to tell her students “Your job, as scientists, is to look for connections!” Ms. Frizzle would love the Mos4-associated Complex (MAC), because MAC has connections all over the place. Indeed, research in mammalian systems and Saccharomyces cerevisiae has shown that MAC, also known as the precursor RNA processing (Prp19) complex, or the NineTeen Complex, functions in splicing, genome stability, protein degradation, and transcription elongation (reviewed in Chanarat and Sträßer, 2013). In plants, the MAC contains orthologs of the components of the Prp19 complex and associates with the spliceosome. The Arabidopsis thaliana MAC contains as core components the MYB transcription factor CELL DIVISION CYCLE5 (CDC5) and the WD40 protein PLEIOTROPIC REGULATORY LOCUS 1 (PRL1), as well as the putative E3 ligase MAC3B, its close homolog MAC3A, and the putative RNA helicase MAC7. In addition to their roles in splicing as part of the MAC, these proteins may also function in microRNA (miRNA) biogenesis. For example CDC5 regulates miRNA gene (MIR) transcription and PRL1 participates in miRNA biogenesis, possibly by stabilizing miRNA precursors (pri-miRNAs) and increasing the activity of DICER-LIKE1 (DCL1).
 To explore miRNA biogenesis in Arabidopsis, Jia et al. (2017) screened for mutants with decreased miRNA activity in plants containing a construct that produces an artificial miRNA targeting CHLORINA42; silencing of CHLORINA42 produces bleaching along the leaf veins and mutants with decreased silencing show less bleaching (de Felippes, Ott, and Weigel, 2011). They identified a mutation in MAC7 as one of the mutants that suppressed silencing. The mac7-1 mutant had pleiotropic root and leaf phenotypes and reduced fertility; a T-DNA insertion mutant, mac7-2, had an embryo-lethal phenotype. Small RNA-sequencing showed reduced levels of 21-nt and 24-nt small RNAs. Consistent with a reduction in miRNA levels, the mac7-1 mutants also had higher mRNA levels for miRNA target genes, including CHLORINA42. The mac7-1 mutants had lower pri-miRNA levels, but the pri-miRNAs had similar half-lives and examination of reporter gene constructs indicated that there was no effect on MIR promoter activity. These findings implicated MAC7 in the promotion of MIR transcription downstream of transcription initiation. The authors also ruled out an indirect effect on the transcription or splicing of miRNA biogenesis factors, showing that the mutants have normal mRNA and protein levels for miRNA-processing factors, including DCL1 and HYPONASTIC LEAVES1 (HYL1).
To explore miRNA biogenesis in Arabidopsis, Jia et al. (2017) screened for mutants with decreased miRNA activity in plants containing a construct that produces an artificial miRNA targeting CHLORINA42; silencing of CHLORINA42 produces bleaching along the leaf veins and mutants with decreased silencing show less bleaching (de Felippes, Ott, and Weigel, 2011). They identified a mutation in MAC7 as one of the mutants that suppressed silencing. The mac7-1 mutant had pleiotropic root and leaf phenotypes and reduced fertility; a T-DNA insertion mutant, mac7-2, had an embryo-lethal phenotype. Small RNA-sequencing showed reduced levels of 21-nt and 24-nt small RNAs. Consistent with a reduction in miRNA levels, the mac7-1 mutants also had higher mRNA levels for miRNA target genes, including CHLORINA42. The mac7-1 mutants had lower pri-miRNA levels, but the pri-miRNAs had similar half-lives and examination of reporter gene constructs indicated that there was no effect on MIR promoter activity. These findings implicated MAC7 in the promotion of MIR transcription downstream of transcription initiation. The authors also ruled out an indirect effect on the transcription or splicing of miRNA biogenesis factors, showing that the mutants have normal mRNA and protein levels for miRNA-processing factors, including DCL1 and HYPONASTIC LEAVES1 (HYL1).
To further tease apart its potential role in miRNA biogenesis, the authors examined MAC7 localization and interactions. Examination of MAC7 fluorescent protein fusions showed that MAC7 localizes to the nucleus, where it overlaps with HYL1 in the nucleoplasm; HYL1 is also enriched in nuclear “dicing bodies” along with DCL1, but MAC7 does not localize to these sub-nuclear structures. The authors next confirmed that MAC7 forms part of the MAC, showing that immunoprecipitation of MAC7 identified a set of proteins that overlapped with published data on proteins immunoprecipitated by MOS4. Bimolecular fluorescence complementation confirmed that MAC7 interacts with CDC5 and (weakly) with PRL1, but MAC7 showed no interaction with HYL1 or DCL1. Although MAC7 and HYL1 did not interact in florescence complementation assays, immunoprecipitation of HYL1 followed by mass spectrometry identified MAC components, including MAC7. This indicates that MAC, as a complex, interacts with the HLY1–DCL1 dicing complex. Moreover, mac7-1 mutants did show reduced localization of HYL1 to dicing bodies; this altered localization may cause the change in small RNA abundance in the mac7-1 mutants.
The authors next explored the role of the MAC in miRNA processing. RNA-sequencing showed that mutants that affected key MAC subunits (mac3a mac3b and prl1 prl2 double mutants) had reduced accumulation of 21-nt and 24-nt small RNAs, consistent with a role for MAC in miRNA biogenesis. Also, the genes that are differentially expressed in these MAC mutants and in the mac7-1 mutants overlapped substantially, indicating that these components function as part of the MAC. Moreover, Gene Ontology analysis found an overrepresentation of functions related to stress responses among the down-regulated genes. Intriguingly, the MAC subunit mutants also accumulated mis-spliced RNAs, showing a high proportion of intron retention; however, genes with mis-spliced transcripts did not show consistent patterns of expression changes in the mutants, indicating separate effects of MAC on gene expression, splicing, and miRNA biogenesis. The specific aspects of MAC function in splicing and miRNA biogenesis remain to be explored. Moreover, how disruption of miRNA processing in the mac7-1 mutant causes a reduction in pri-miRNA levels remains to be clarified.
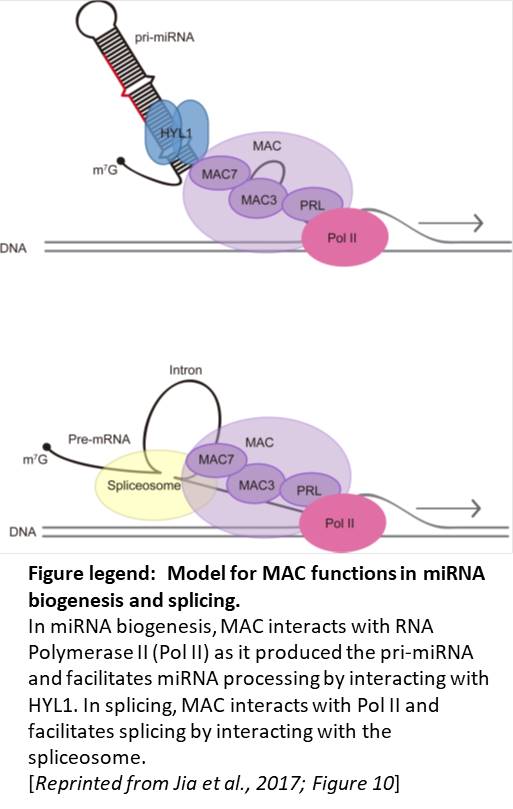
In sum, this study firmly connects MAC to yet another aspect of RNA metabolism, miRNA biogenesis, indicating that MAC may function to connect transcription with RNA processing in mRNA splicing and in miRNA biogenesis (see figure). Ms. Frizzle would be proud.
REFERENCES
Chanarat S. and Sträßer K. (2013) Splicing and beyond: the many faces of the Prp19 complex. Biochim Biophys Acta. 1833(10): 2126-34 doi: 10.1016/j.bbamcr.2013.05.023
de Felippes, F.F., Ott, F., and Weigel, D. (2011). Comparative analysis of non-autonomous effects of tasiRNAs and miRNAs in Arabidopsis thaliana. Nucleic Acids Res 39, 2880-2889.
Jia, T., Zhang, B., You, C., Zhang, Y., Zeng, L., Li, S., Johnson, K.C.M. Yu, B., Li, X., and Chen, X. (2017) The Arabidopsis MOS4-associated Complex Promotes MicroRNA Biogenesis and Precursor Messenger RNA Splicing. Plant Cell 10.1105/tpc.17.00370.



