Chloroplasts Use Bacterial Mechanism to Recognize Start Codons
Scharff et al. investigate function of anti-Shine-Dalgarno sequence in chloroplast ribosomes https://doi.org/10.1105/tpc.17.00524
By Lars B. Scharff
BACKGROUND: Cells of plants contain three different systems for protein synthesis: in the cytosol, in plastids, and in mitochondria. Proteins in the cytosol are made by typical eukaryotic ribosomes. According to the endosymbiotic theory, plastids and mitochondria have bacterial ancestors and their ribosomes are similar to bacterial ribosomes. Previously there was no agreement on the mechanisms of how plastid ribosomes recognize the start codons. It was not clear if the mechanisms are similar to the ones of the bacterial ancestors of plastids or if completely different mechanisms have evolved during the integration of the former bacterial cell into the metabolism and regulation of the eukaryotic plant cell.
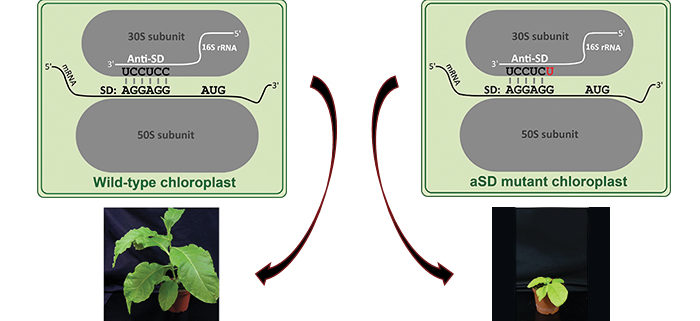
QUESTIONS: One of the potential bacterial mechanisms is based on binding of the ribosome via the so-called anti-Shine-Dalgarno sequence (aSD) to a sequence on the mRNA called the Shine-Dalgarno sequence (SD). We mutated the ribosomal aSD in plastids to test if this mechanism is used for start codon recognition in plastids.
FINDINGS: The aSD mutants in tobacco (Nicotiana tabacum) show that this mechanism is active in plastids. We produced plants with different mutations. Milder mutations caused reduced growth, whereas we could not obtain the stronger mutants indicating that tobacco could not survive with those. We analyzed protein synthesis in the mutant with the mildest phenotype by measuring how many ribosomes bind to mRNA. The translation of mRNAs that have SDs and were predicted to be impaired by the mutation was reduced in the aSD mutant. By contrast, the translation of mRNAs lacking SDs was not influenced by the aSD mutation. Interestingly, mRNAs with SDs and with start codons exposed in their mRNA secondary structure were less dependent on the SD, indicating a role of the structure.
NEXT STEPS: Future research is aimed at how protein synthesis in plastids is adjusted to the needs of plants. How can plants influence the recognition of start codons to increase or decrease the translation of a specific gene? How can translation of a specific gene can be switched on or switch off completely?
Lars B Scharff, Miriam Ehrnthaler, Marcin Janowski, Liam H Childs, Claudia Hasse, Jürgen Gremmels, Stephanie Ruf, Reimo Zoschke, Ralph Bock. (2017) Shine-Dalgarno Sequences Play an Essential Role in the Translation of Plastid mRNAs in Tobacco. Plant Cell. Published November 2017. DOI: https://doi.org/10.1105/tpc.17.00524