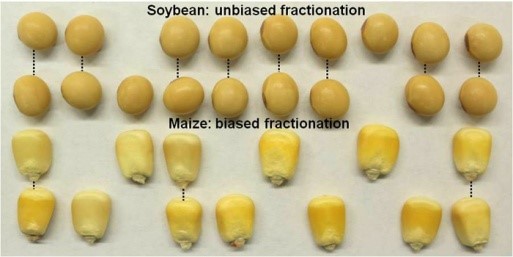
Duplicate Genomes Evolved Differently in Maize and Soybean
Zhao et al. demonstrate that duplicated genomes in maize and soybean followed distinct trajectories over millions of years https://doi.org/10.1105/tpc.17.00595
By Meixia Zhao, Biao Zhang, Damon Lisch, and Jianxin Ma
Background: Over evolutionary time, many organisms, particularly plants, have periodically experienced whole-genome duplication. This can happen spontaneously within a species, or occur during mating between related species. As a consequence, each plant will carry two subgenomes, and all genes will have a duplicate copy. For many of these paired genes, functional redundancy results in the loss of one copy, a process known as fractionation. Many plant species show biased fractionation, whereby one subgenome experienced more gene loss than the other. The cause of this unclear, but transposable elements or “jumping genes” have been implicated, due to the repressive effects they can have on nearby genes.
Questions: Soybean and maize each experienced whole-genome duplication events that happened at roughly the same time. What are the similarities and differences in subgenome differentiation between maize and soybean? What are the potential causes for subgenome differentiation?
Findings: We observed distinct patterns of gene and genome differentiation when comparing these two species. In agreement with previous work, we found that one of the two maize subgenomes had higher rates of gene loss, lower average levels of gene expression, higher rates of transposable element accumulation, and more small interfering RNAs and DNA methylation near its genes. In contrast, we did not observe any of these differences between the subgenomes of soybeans. Individual gene pairs exhibit differentiation in soybean, but the process is gene-by-gene, rather than genome-by-genome. Soybean also has much lower frequency of gene loss and more chromosomal rearrangements than maize. We hypothesize that whole-genome duplication results in a competition between paired genes, and that in maize, but not in soybean, one subgenome began with a disadvantage, which resulted in higher rates of fractionation. By contrast, the two subgenomes of soybean were much more similar to each other when the whole-genome duplication event occurred, resulting a lack of bias and a higher frequency of gene retention.
Next steps: A primary question is if transposable elements are a cause or a consequence of subgenome differentiation. We are also interested in the functional consequences of the retention of so many more duplicate genes in soybean relative to maize.
Zhao, M., Zhang, B., Lisch, D. and Ma, J. (2017). Patterns and Consequences of Subgenome Differentiation Provide Insights into the Nature of Paleopolyploidy in Plants. Plant Cell https://doi.org/10.1105/tpc.17.00595