A molecular toolkit for screening elite rhizobia (PNAS)
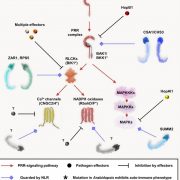
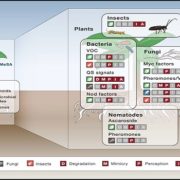
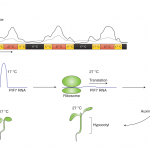
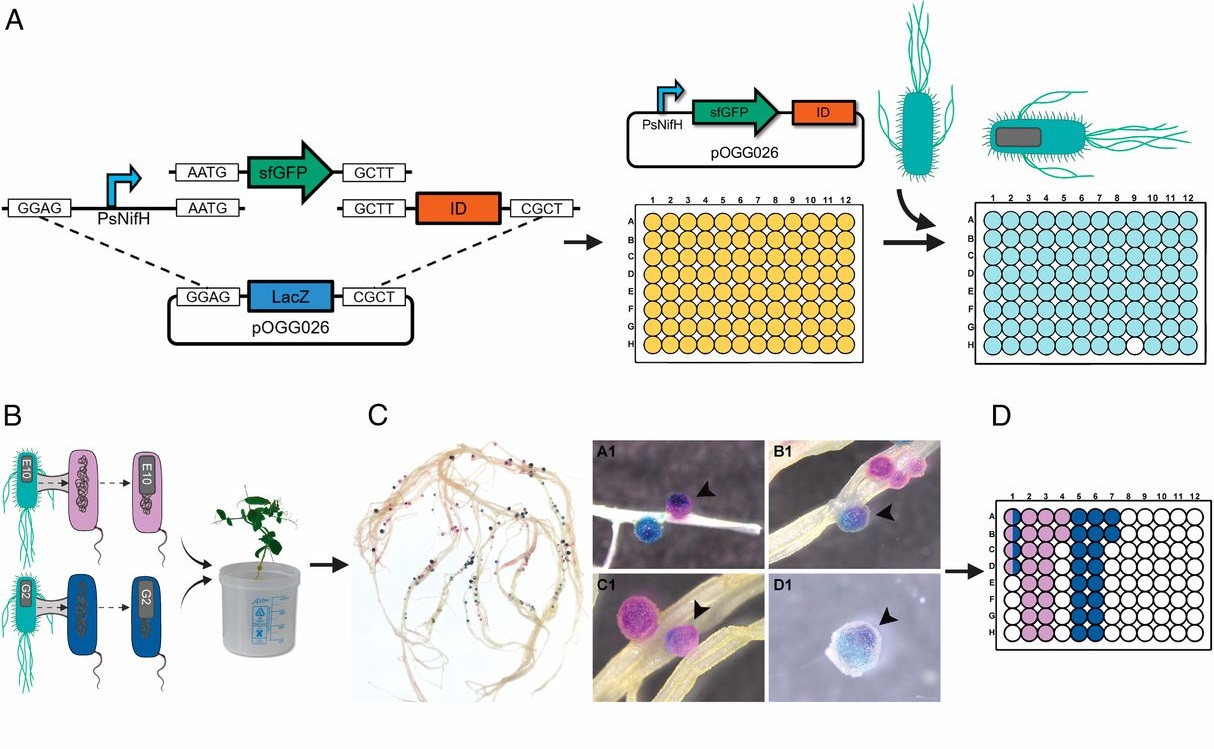 N2-fixing rhizobia bacteria are able to establish symbiotic interactions with legumes in specialized organs called root nodules. Identifying elite rhizobia that are both competitive for nodule occupancy and effective in N2 fixation in agricultural environments is crucial for maximizing the yield of legumes. Mendoza-Suárez et al. developed a high-throughput strategy for simultaneously assessing competitiveness and effectiveness of rhizobia strains in plants. The authors generated a plasmid library containing a synthetic nifH promoter-driven superfolder GFP, which reports symbiosis-induced nitrogenase activity, and unique barcodes, which can be sequenced for quantifying bacterial population. 84 Rhizobium leguminosarum strains conjugated with the uniquely barcoded plasmids were inoculated into pea plants grown in natural soil and screened for their abundance and nitrogenase activity. This screening identified G083 as the most competitive and highly efficient strain, and the authors validated that G083 can enhance plant performance better than other strains. Interestingly, the screening found many nodules occupied with multiple rhizobia strains, suggesting that complex interspecies interactions may exist inside nodules. The tool developed in this study allows not only a high-throughput and cost-efficient screening of elite rhizobia strains but also molecular ecological analysis of rhizobia in the context of the plant microbiota. (Summary by Tatsuya Nobori @nobolly) PNAS10.1073/pnas.1921225117
N2-fixing rhizobia bacteria are able to establish symbiotic interactions with legumes in specialized organs called root nodules. Identifying elite rhizobia that are both competitive for nodule occupancy and effective in N2 fixation in agricultural environments is crucial for maximizing the yield of legumes. Mendoza-Suárez et al. developed a high-throughput strategy for simultaneously assessing competitiveness and effectiveness of rhizobia strains in plants. The authors generated a plasmid library containing a synthetic nifH promoter-driven superfolder GFP, which reports symbiosis-induced nitrogenase activity, and unique barcodes, which can be sequenced for quantifying bacterial population. 84 Rhizobium leguminosarum strains conjugated with the uniquely barcoded plasmids were inoculated into pea plants grown in natural soil and screened for their abundance and nitrogenase activity. This screening identified G083 as the most competitive and highly efficient strain, and the authors validated that G083 can enhance plant performance better than other strains. Interestingly, the screening found many nodules occupied with multiple rhizobia strains, suggesting that complex interspecies interactions may exist inside nodules. The tool developed in this study allows not only a high-throughput and cost-efficient screening of elite rhizobia strains but also molecular ecological analysis of rhizobia in the context of the plant microbiota. (Summary by Tatsuya Nobori @nobolly) PNAS10.1073/pnas.1921225117
[altmetric doi=”10.1073/pnas.1921225117″ details=”right” float=”right”]