Genetic Basis of Natural Variation of Rice Ionomics
(Translated from the Chinese original http://news.hzau.edu.cn/2018/1101/52991.shtml)
Nanhu News Network (Correspondent Sheng Ke)
On October 30th, the research group of Professor Tian Xingming from the Rice Science Team of the National Key Laboratory of Crop Genetic Improvement of our School of Life Science and Technology, published in the Plant Cell, “Genetic basis of rice ionomic variation revealed by Genome-wide association studies“.
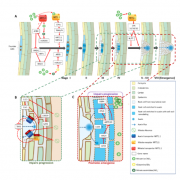
The study revealed the genetic basis for the natural variation of the rice ionome through genome-wide association analysis. Plants require at least 14 essential mineral nutrients and multiple beneficial elements for growth, development, and resistance to biotic and abiotic stresses. At the same time, the soil also contains some toxic elements, which are not essential for the growth and development of plants. Plants generally do not actively absorb them, but the toxic elements can use the ion channels of essential elements to enter the plant, which may lead to plant poisoning or threats entering the food chain and affecting human health. For people who eat rice as the main food, rice is the main dietary source of many important micronutrients as well as toxic elements, so increasing the accumulation of important micronutrients in rice and reducing the concentration of potentially toxic elements is critical to food safety and human health. Past studies have shown that different rice varieties have great differences in the accumulation of mineral elements, but the genetic basis for controlling the natural variation of rice ionomics still largely unknown. Therefore, systematic discovery of the corresponding genetic loci is conducive to the genetic improvement of rice nutritional quality.
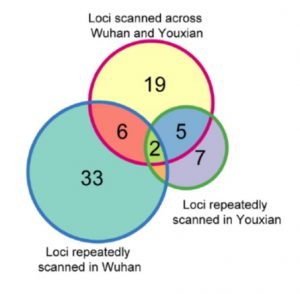 The research team used 529 rice germplasm materials to conduct multi-year field trials in Hubei and Hunan, and determined the content of 17 mineral elements in straw and grain, combined with rice genome-wide single base sequence polymorphism. The correlation analysis of the information analyzed the genetic basis of the natural variation of the rice ionomics. In the correlation analysis of 17 elemental traits, 72 highly reproducible sites were found, of which 32 were repeatedly detected at different test sites, and 40 sites were in different fields at the same test site. In mining of candidate genes, the authors predicted candidate genes at 42 loci and functionally verified the candidate genes of three loci, including OsHKT1; 5, OsMOT1; 1 and Ghd7. OsHKT1;5 is a known sodium transporter, and its haplotype analysis further confirms its functional variation in natural populations. OsMOT1;1 is a homologous gene of Arabidopsis molybdenum transporter AtMOT1 The transgenic method was used to confirm the molybdenum transport function and natural variation mechanism in rice. Ghd7 is a known heading and yield trait gene, which was confirmed by near isogenic lines and transgenic materials to affect the nitrogen content in rice plants. This study provides useful genetic loci and theoretical bases for the genetic improvement of rice nutritional quality.
The research team used 529 rice germplasm materials to conduct multi-year field trials in Hubei and Hunan, and determined the content of 17 mineral elements in straw and grain, combined with rice genome-wide single base sequence polymorphism. The correlation analysis of the information analyzed the genetic basis of the natural variation of the rice ionomics. In the correlation analysis of 17 elemental traits, 72 highly reproducible sites were found, of which 32 were repeatedly detected at different test sites, and 40 sites were in different fields at the same test site. In mining of candidate genes, the authors predicted candidate genes at 42 loci and functionally verified the candidate genes of three loci, including OsHKT1; 5, OsMOT1; 1 and Ghd7. OsHKT1;5 is a known sodium transporter, and its haplotype analysis further confirms its functional variation in natural populations. OsMOT1;1 is a homologous gene of Arabidopsis molybdenum transporter AtMOT1 The transgenic method was used to confirm the molybdenum transport function and natural variation mechanism in rice. Ghd7 is a known heading and yield trait gene, which was confirmed by near isogenic lines and transgenic materials to affect the nitrogen content in rice plants. This study provides useful genetic loci and theoretical bases for the genetic improvement of rice nutritional quality.
Dr. Yang Xing, a postdoctoral fellow of Professor Xing Xingming, and Lu Kai, a Ph.D. student, are the co-first authors of the paper, and Professor Xing Xingming is the author of the communication. The research was funded by the National Natural Science Foundation, the 863 Program, and the Public Welfare Industry (Agriculture) Research Program.
Article link:
Http://www.plantcell.org/content/early/2018/10/29/tpc.18.00375
Reviewer: Lian Xingming