ARGONAUTE1: To Shed a Strand or Not?
Clavel et al. investigate recognition and binding of AGO1 by viral P0 protein https://doi.org/10.1105/tpc.18.00111.
By Marion Clavel
Background: In plants, small RNAs (sRNAs) are widely used to regulate gene expression of both host genes and foreign nucleic acid, like viruses. sRNAs function in this way by associating with an effector protein called ARGONAUTE (AGO). sRNAs are produced as a duplex of two complementary strands. The AGO protein acts by “loading” the sRNA duplex, then removing one of the two strands and forming an RNA-Induced Silencing Complex (RISC). The sRNA strand that remains acts as a template for RISC to recognize and cleave a complementary target RNA (mRNA transcript or viral RNA). To protect itself from this mechanism, the Turnip yellow virus produces the P0 protein that degrades the AGO1 protein; but how P0 recognizes AGO1 is not yet known.
Question: We wanted to know how P0 recognizes AGO1 and what host factors, if any, does it use to achieve this targeting. Using the model plant Arabidopsis thaliana, we employed two complementary approaches to help pinpoint amino acids in AGO1 that are important for P0 recognition.
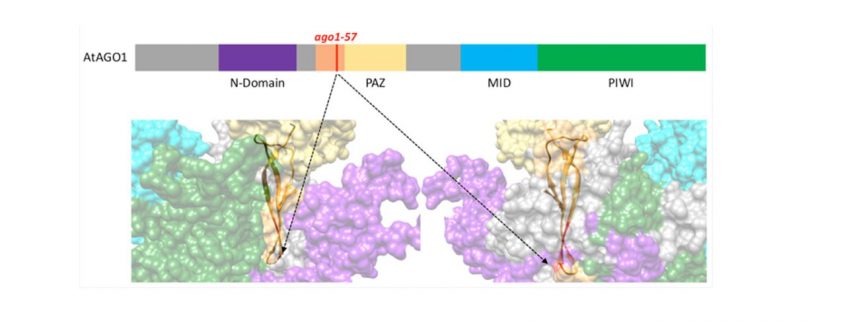
Findings: We found that mutating one particular amino acid of AGO1 is enough to block the binding of P0 to AGO1, and therefore its degradation. Intriguingly, this particular site is not directly accessible on the surface of the protein, suggesting that AGO1 adopts different conformations, one of which is preferred by P0. Moreover, we found that the uncharacterized domain in which this amino acid is found is important for AGO1 to “shed” the complementary strand of the sRNA duplex, and thus is an intrinsic part of making a functional RISC. Our study also highlights how AGO1 copes with two different types of sRNAs: Small interfering RNA duplexes that are fully complementary are not processed by the mutated AGO1, while most microRNA duplexes that are not perfectly complementary are mostly unaffected.
Next steps: We are now working on identifying other proteins that might be important for mediating the degradation of AGO1 by P0. We are also investigating the importance of the mutated AGO1 protein in the context of infection by Turnip yellow Virus and other plant viruses.
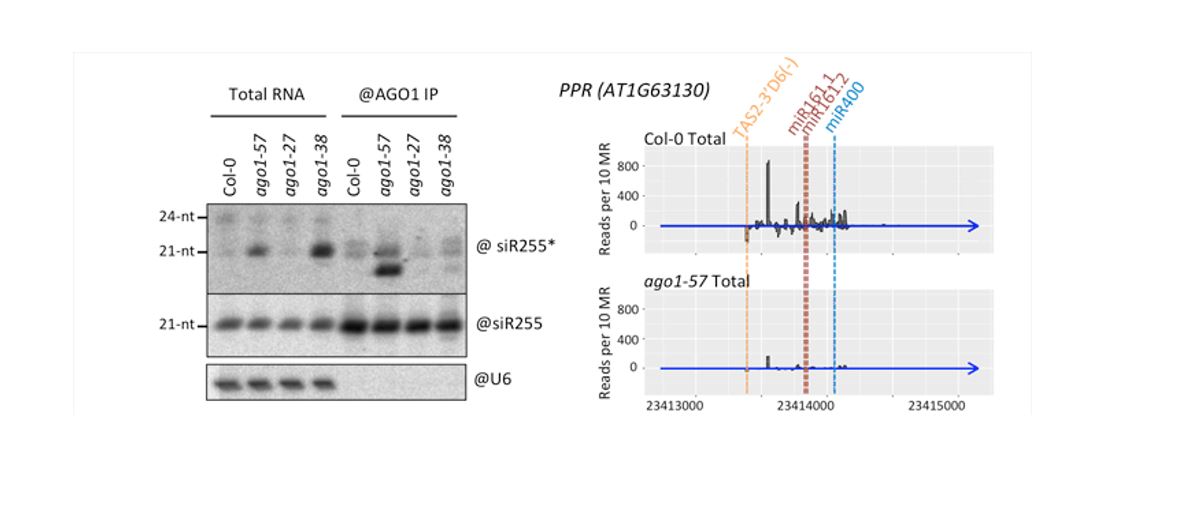
Derrien, Clavel, et al. (2018). A suppressor screen for AGO1 degradation by the viral F-Box P0 protein uncovers a role for AGO DUF1785 in sRNA duplex unwinding. Plant Cell https://doi.org/10.1105/tpc.18.00111.
Key words: RNA silencing, SCF E3 ligase, viral suppressor of silencing, secondary small RNAs.