Plant Science Research Weekly: May 8th
Tissue-resolved multi-omics atlas of Arabidopsis
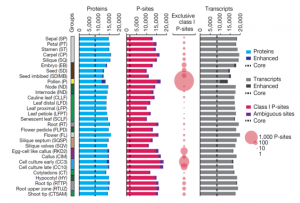 Arabidopsis as a model system has been intensively studied over the past twenty years, but the proteome of Arabidopsis has been less well characterized. Here, Mergner et al. report the first 30-tissue-type integrated proteome, phosphoproteome and transcriptome atlas of Arabidopsis. The data cover the abundance of more than 18,000 proteins out of more than 27,000 annotated protein-coding genes, and identifies nearly 44,000 phosphorylation sites (P-sites), making this one of the most extensive studies so far. In general, these findings show a positive correlation between transcript and protein level in almost all the tissue types. Mass spectrometry analysis revealed spice-site variants and peptides translated from short open-reading frames, demonstrating the power of these tools to improve genome annotation. The study detected about 700 kinases and 120 phosphatases across most of the tissues. By motif-X algorithm, this study identifies 266 phosphorylation motifs grouped into different classes, and the most complete Arabidopsis phosphoproteome to date. Overall, this study presents a deeply analysed multi-omics data set. As an example of its power, the authors show how it can reveal tissue-specific complexes or motif-based phosphorylation events. These data can be accessed by using web portals ATHENA and ProteomicsDB. (Summary by Min May Wong @wongminmay) Nature 10.1038/s41586-020-2094-2
Arabidopsis as a model system has been intensively studied over the past twenty years, but the proteome of Arabidopsis has been less well characterized. Here, Mergner et al. report the first 30-tissue-type integrated proteome, phosphoproteome and transcriptome atlas of Arabidopsis. The data cover the abundance of more than 18,000 proteins out of more than 27,000 annotated protein-coding genes, and identifies nearly 44,000 phosphorylation sites (P-sites), making this one of the most extensive studies so far. In general, these findings show a positive correlation between transcript and protein level in almost all the tissue types. Mass spectrometry analysis revealed spice-site variants and peptides translated from short open-reading frames, demonstrating the power of these tools to improve genome annotation. The study detected about 700 kinases and 120 phosphatases across most of the tissues. By motif-X algorithm, this study identifies 266 phosphorylation motifs grouped into different classes, and the most complete Arabidopsis phosphoproteome to date. Overall, this study presents a deeply analysed multi-omics data set. As an example of its power, the authors show how it can reveal tissue-specific complexes or motif-based phosphorylation events. These data can be accessed by using web portals ATHENA and ProteomicsDB. (Summary by Min May Wong @wongminmay) Nature 10.1038/s41586-020-2094-2
Plant 22-nt siRNAs mediate translational repression and stress adaptation
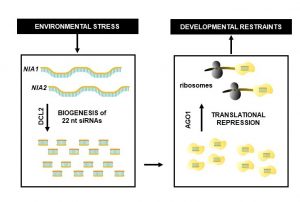 Among the myriad small interfering RNAs, 21- and 24-nucleotides siRNAs control plant development and immunity through mRNA cleavage and RNA-directed DNA methylation, respectively. Still, the regulation and biological function of 22-nt siRNAs remain unresolved. In this report, Wu and coworkers investigated the massive accumulation of 22-nt siRNAs in plants with mutations in DICER-like 4 (DCL4) and RNA-degrading enzymes ETHYLENE INSENSITIVE5 (EIN5) and SUPER KILLER2 (SKI2), and found that two genes encoding nitrate reductases (NIA1, NIA2) contributed to almost half of the total amount of 22-nt siRNAs. Noteworthy, transcriptome analysis and polysome profiling of NIA1/2 revealed that 22-nt siRNAs repress translation globally as well as gene-specifically. Plants that are blocked in RNA decay and accumulate 22-nt siRNAs (ein5 dcl4 and ski2 dcl4) display root growth inhibition, abnormal phloem development and anthocyanin accumulation. Similar pleiotropic defects and accumulation of 22-nt siRNAs were also observed in wild-type plants in response to abiotic stress (i.e., nitrogen starvation, high salinity and ABA treatment), indicating a key role for 22-nt siRNAs in the fine-tuning of stress adaptation. To conclude, stress leading to an increase in 22-nt siRNAs likely causes a shortage of amino acids mediated by translational repression of NIA1/2, and consequently the slowdown of global translation leading to developmental restraint. Thus, this strategy allows plants to switch from growth to defense under environmental limitations. (Summary by Michela Osnato @michela_osnato) Nature 10.1038/s41586-020-2231-y
Among the myriad small interfering RNAs, 21- and 24-nucleotides siRNAs control plant development and immunity through mRNA cleavage and RNA-directed DNA methylation, respectively. Still, the regulation and biological function of 22-nt siRNAs remain unresolved. In this report, Wu and coworkers investigated the massive accumulation of 22-nt siRNAs in plants with mutations in DICER-like 4 (DCL4) and RNA-degrading enzymes ETHYLENE INSENSITIVE5 (EIN5) and SUPER KILLER2 (SKI2), and found that two genes encoding nitrate reductases (NIA1, NIA2) contributed to almost half of the total amount of 22-nt siRNAs. Noteworthy, transcriptome analysis and polysome profiling of NIA1/2 revealed that 22-nt siRNAs repress translation globally as well as gene-specifically. Plants that are blocked in RNA decay and accumulate 22-nt siRNAs (ein5 dcl4 and ski2 dcl4) display root growth inhibition, abnormal phloem development and anthocyanin accumulation. Similar pleiotropic defects and accumulation of 22-nt siRNAs were also observed in wild-type plants in response to abiotic stress (i.e., nitrogen starvation, high salinity and ABA treatment), indicating a key role for 22-nt siRNAs in the fine-tuning of stress adaptation. To conclude, stress leading to an increase in 22-nt siRNAs likely causes a shortage of amino acids mediated by translational repression of NIA1/2, and consequently the slowdown of global translation leading to developmental restraint. Thus, this strategy allows plants to switch from growth to defense under environmental limitations. (Summary by Michela Osnato @michela_osnato) Nature 10.1038/s41586-020-2231-y
An RNA thermoswitch regulates daytime growth in Arabidopsis
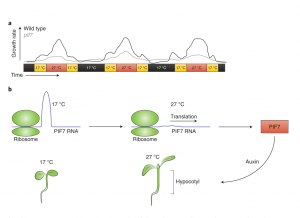 Plants are sessile organisms and as such they must be highly responsive to environmental cues including temperature, and adjust their growth and development accordingly. Considering that mean global surface temperatures are expected to continue to rise dramatically in the 21st century, it is crucial to understand how plants respond to elevated temperatures. Here, Chung et al. reveal that the mRNA of the transcription factor PHYTOCHROME INTERACTING FACTOR7 (PIF7) behaves like an “RNA thermometer” to regulate daytime growth in Arabidopsis (check out this twitter thread). Using ribosome profiling and RNA-Seq, the authors found that PIF7 was preferentially translated in response to warmer daytime temperatures. A vast array of techniques were employed to uncover the underlying mechanism behind this phenomenon. It was revealed that an RNA hairpin exists within the 5′-untranslated region of PIF7 which at lower temperatures (17°C) prevents translation. However, warmer temperatures (27°C) lead to a partial unfolding of the hairpin, triggering a more relaxed conformation, facilitating translation. Elevated PIF7 translation induces the transcription of genes required for growth and auxin biosynthesis and mutagenesis experiments demonstrated the pronounced role of PIF7 in thermomorphogenesis, namely regarding hypocotyl elongation and stomata formation. Furthermore, similar RNA thermoswitch mechanisms were revealed for the transcription factors WRKY22 and HSFA2. Thus, RNA thermoswitches may represent a conserved mechanism for adaptive responses to high temperatures in plants, symbolizing an exciting prospect for the plant thermomorphogenesis research field. (Summary by Caroline Dowling @CarolineD0wling ) Nature Plants 10.1038/s41477-020-0633-3 (Image credit: Kasper van Gelderen and Ronald Pierik 10.1038/s41477-020-0643-1)
Plants are sessile organisms and as such they must be highly responsive to environmental cues including temperature, and adjust their growth and development accordingly. Considering that mean global surface temperatures are expected to continue to rise dramatically in the 21st century, it is crucial to understand how plants respond to elevated temperatures. Here, Chung et al. reveal that the mRNA of the transcription factor PHYTOCHROME INTERACTING FACTOR7 (PIF7) behaves like an “RNA thermometer” to regulate daytime growth in Arabidopsis (check out this twitter thread). Using ribosome profiling and RNA-Seq, the authors found that PIF7 was preferentially translated in response to warmer daytime temperatures. A vast array of techniques were employed to uncover the underlying mechanism behind this phenomenon. It was revealed that an RNA hairpin exists within the 5′-untranslated region of PIF7 which at lower temperatures (17°C) prevents translation. However, warmer temperatures (27°C) lead to a partial unfolding of the hairpin, triggering a more relaxed conformation, facilitating translation. Elevated PIF7 translation induces the transcription of genes required for growth and auxin biosynthesis and mutagenesis experiments demonstrated the pronounced role of PIF7 in thermomorphogenesis, namely regarding hypocotyl elongation and stomata formation. Furthermore, similar RNA thermoswitch mechanisms were revealed for the transcription factors WRKY22 and HSFA2. Thus, RNA thermoswitches may represent a conserved mechanism for adaptive responses to high temperatures in plants, symbolizing an exciting prospect for the plant thermomorphogenesis research field. (Summary by Caroline Dowling @CarolineD0wling ) Nature Plants 10.1038/s41477-020-0633-3 (Image credit: Kasper van Gelderen and Ronald Pierik 10.1038/s41477-020-0643-1)
MASS proteins and stomatal development in Arabidopsis
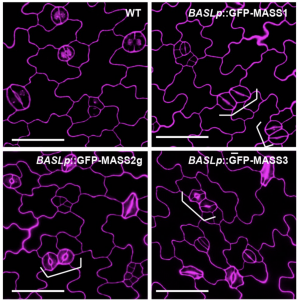 Stomata are the pores surrounded by a pair of guard cells on the plant epidermis that help in gaseous exchange. The number and spacing of stomata are regulated by a series of phosphorylation and de-phosphorylation events of key transcription factors through the mitogen-activated protein kinase (MAPK) signal cascade [which includes MAPK kinases (MAPKKs), and MAPKK kinases (MAPKKKs)]. One of the well-known pathways in stomatal development is a series of phosphorylation events by YODA (a MAPKKK), MAPKK4/5, and MAPK 3/6, leading to the eventual inhibition of SPEECHLESS, transcription factor involved in stomatal development. In this paper Xue et al. characterized three other substrates of MAPK signaling that they named MAPK SUBSTRATES IN THE STOMATAL LINEAGE (MASS) proteins. Using biochemical and genetic experiments, the authors showed that MASS proteins regulate the number and spacing between adjacent stomata. Further, mechanistically MASS proteins are localized to the plasma membrane and interact with YODA to negatively regulate the MAPK signaling. Thus, this paper advances our understanding of stomatal development by adding three more MASS proteins in the stomatal developmental pathway. (Summary by Vijaya Batthula @Vijaya_Batthula). PLOS Genetics 10.1371/journal.pgen.1008706
Stomata are the pores surrounded by a pair of guard cells on the plant epidermis that help in gaseous exchange. The number and spacing of stomata are regulated by a series of phosphorylation and de-phosphorylation events of key transcription factors through the mitogen-activated protein kinase (MAPK) signal cascade [which includes MAPK kinases (MAPKKs), and MAPKK kinases (MAPKKKs)]. One of the well-known pathways in stomatal development is a series of phosphorylation events by YODA (a MAPKKK), MAPKK4/5, and MAPK 3/6, leading to the eventual inhibition of SPEECHLESS, transcription factor involved in stomatal development. In this paper Xue et al. characterized three other substrates of MAPK signaling that they named MAPK SUBSTRATES IN THE STOMATAL LINEAGE (MASS) proteins. Using biochemical and genetic experiments, the authors showed that MASS proteins regulate the number and spacing between adjacent stomata. Further, mechanistically MASS proteins are localized to the plasma membrane and interact with YODA to negatively regulate the MAPK signaling. Thus, this paper advances our understanding of stomatal development by adding three more MASS proteins in the stomatal developmental pathway. (Summary by Vijaya Batthula @Vijaya_Batthula). PLOS Genetics 10.1371/journal.pgen.1008706
Intertwined signatures of desiccation and drought tolerance in grasses
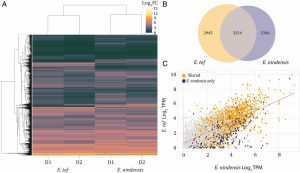 Desiccation tolerance (i.e., the capacity of surviving with very low water content) is widespread in seeds and pollen, but quite rare in vegetative organs. Most authors agree that in angiosperms it originated multiple independent times from rewiring seed desiccation tolerance pathways. Here, Pardo et al. sequenced the genome of Eragrostis nindensis, a species from a genus that includes both desiccation-tolerant and -sensitive species and crops, and compared gene expression under water deficit with other desiccation-tolerant and -sensitive grass species. Interestingly, the authors found seed-related genes upregulated in both desiccation-tolerant and -sensitive species under drought. As a result, they challenge the current hypothesis for desiccation tolerance origin and suggest that seed-related genes, rather than participating only in desiccation tolerance responses, are involved in general drought tolerance responses. However, the genes related to preventing photooxidative damage had different expression levels between desiccation-tolerant and desiccation-sensitive species. Since a role for photooxidative damage in desiccation tolerance has been previously reported, the authors suggest these genes are crucial for the evolution of vegetative desiccation-tolerance. Moreover, they consider these findings are promising for future efforts to improve stress resilience in crops (Summary by Carlos A. Ordóñez-Parra @caordonezparra) PNAS 10.1073/pnas.2001928117
Desiccation tolerance (i.e., the capacity of surviving with very low water content) is widespread in seeds and pollen, but quite rare in vegetative organs. Most authors agree that in angiosperms it originated multiple independent times from rewiring seed desiccation tolerance pathways. Here, Pardo et al. sequenced the genome of Eragrostis nindensis, a species from a genus that includes both desiccation-tolerant and -sensitive species and crops, and compared gene expression under water deficit with other desiccation-tolerant and -sensitive grass species. Interestingly, the authors found seed-related genes upregulated in both desiccation-tolerant and -sensitive species under drought. As a result, they challenge the current hypothesis for desiccation tolerance origin and suggest that seed-related genes, rather than participating only in desiccation tolerance responses, are involved in general drought tolerance responses. However, the genes related to preventing photooxidative damage had different expression levels between desiccation-tolerant and desiccation-sensitive species. Since a role for photooxidative damage in desiccation tolerance has been previously reported, the authors suggest these genes are crucial for the evolution of vegetative desiccation-tolerance. Moreover, they consider these findings are promising for future efforts to improve stress resilience in crops (Summary by Carlos A. Ordóñez-Parra @caordonezparra) PNAS 10.1073/pnas.2001928117
Mechanical shielding in plant nuclei
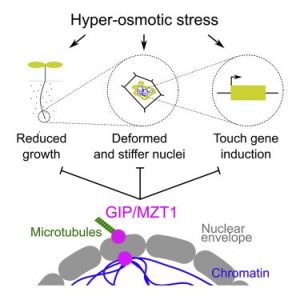 The nucleus is an organelle with tremendous shape flexibility in response to environmental cues; it has been described as the “plastic, elastic, and fantastic” organelle. The change in nuclear geometry based on mechanical stress is well documented from single cell studies in culture, but the question remains of how the nuclear geometry alters due to mechanical cues in a multicellular organism. Goswami et al. addressed this question using Arabidopsis roots under hyperosmotic stress. Hyperosmotic stress reduces root growth, leads to deformed and stiff nuclei, and eventually induces the expression of touch-related genes. Reversing the osmotic stress, either through iso-osmotic or hypo-osmotic conditions, reverses these effects. Previously, the authors showed that GIP gamma-tubulin complex-associated proteins are involved in nuclear architecture in Arabidopsis. Here, they found that in the gip1gip2 mutant, nuclei resemble those of cells experiencing hyperosmotic stress and the mutants constitutively appear to be under hyperosmotic stress. The mutant also shows enhanced growth and survival as compared to the wild-type when under hyperosmotic stress. These findings raise the question of how osmolarity affects nuclear shape, and opens the door to new opportunities to investigate drought responses in plants. (Summary by Arif Ashraf) Current Biology 10.1016/j.cub.2020.03.059
The nucleus is an organelle with tremendous shape flexibility in response to environmental cues; it has been described as the “plastic, elastic, and fantastic” organelle. The change in nuclear geometry based on mechanical stress is well documented from single cell studies in culture, but the question remains of how the nuclear geometry alters due to mechanical cues in a multicellular organism. Goswami et al. addressed this question using Arabidopsis roots under hyperosmotic stress. Hyperosmotic stress reduces root growth, leads to deformed and stiff nuclei, and eventually induces the expression of touch-related genes. Reversing the osmotic stress, either through iso-osmotic or hypo-osmotic conditions, reverses these effects. Previously, the authors showed that GIP gamma-tubulin complex-associated proteins are involved in nuclear architecture in Arabidopsis. Here, they found that in the gip1gip2 mutant, nuclei resemble those of cells experiencing hyperosmotic stress and the mutants constitutively appear to be under hyperosmotic stress. The mutant also shows enhanced growth and survival as compared to the wild-type when under hyperosmotic stress. These findings raise the question of how osmolarity affects nuclear shape, and opens the door to new opportunities to investigate drought responses in plants. (Summary by Arif Ashraf) Current Biology 10.1016/j.cub.2020.03.059
A molecular toolkit for screening elite rhizobia
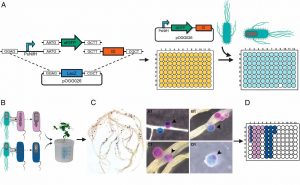 N2-fixing rhizobia bacteria are able to establish symbiotic interactions with legumes in specialized organs called root nodules. Identifying elite rhizobia that are both competitive for nodule occupancy and effective in N2 fixation in agricultural environments is crucial for maximizing the yield of legumes. Mendoza-Suárez et al. developed a high-throughput strategy for simultaneously assessing competitiveness and effectiveness of rhizobia strains in plants. The authors generated a plasmid library containing a synthetic nifH promoter-driven superfolder GFP, which reports symbiosis-induced nitrogenase activity, and unique barcodes, which can be sequenced for quantifying bacterial population. 84 Rhizobium leguminosarum strains conjugated with the uniquely barcoded plasmids were inoculated into pea plants grown in natural soil and screened for their abundance and nitrogenase activity. This screening identified G083 as the most competitive and highly efficient strain, and the authors validated that G083 can enhance plant performance better than other strains. Interestingly, the screening found many nodules occupied with multiple rhizobia strains, suggesting that complex interspecies interactions may exist inside nodules. The tool developed in this study allows not only a high-throughput and cost-efficient screening of elite rhizobia strains but also molecular ecological analysis of rhizobia in the context of the plant microbiota. (Summary by Tatsuya Nobori @nobolly) PNAS10.1073/pnas.1921225117
N2-fixing rhizobia bacteria are able to establish symbiotic interactions with legumes in specialized organs called root nodules. Identifying elite rhizobia that are both competitive for nodule occupancy and effective in N2 fixation in agricultural environments is crucial for maximizing the yield of legumes. Mendoza-Suárez et al. developed a high-throughput strategy for simultaneously assessing competitiveness and effectiveness of rhizobia strains in plants. The authors generated a plasmid library containing a synthetic nifH promoter-driven superfolder GFP, which reports symbiosis-induced nitrogenase activity, and unique barcodes, which can be sequenced for quantifying bacterial population. 84 Rhizobium leguminosarum strains conjugated with the uniquely barcoded plasmids were inoculated into pea plants grown in natural soil and screened for their abundance and nitrogenase activity. This screening identified G083 as the most competitive and highly efficient strain, and the authors validated that G083 can enhance plant performance better than other strains. Interestingly, the screening found many nodules occupied with multiple rhizobia strains, suggesting that complex interspecies interactions may exist inside nodules. The tool developed in this study allows not only a high-throughput and cost-efficient screening of elite rhizobia strains but also molecular ecological analysis of rhizobia in the context of the plant microbiota. (Summary by Tatsuya Nobori @nobolly) PNAS10.1073/pnas.1921225117