Life is Sweeter with Trehalose 6-Phosphate
Besides making our daily lives sweeter, sugars are the most important energy source in all organisms. Balancing their availability and regulating their partitioning between tissues are major determinants for growth and development. In plants, sucrose is the main product of photosynthesis and is transported from source to sink organs through the phloem. Sucrose levels are regulated by the disaccharide trehalose 6-phosphate (Tre6P) which also acts as a signaling metabolite coordinating sugar status, carbon assimilation and development (Lunn et al., 2014).
Tre6P is synthesized from two glucose moieties by Trehalose-6-Phosphate Synthase 1 (TPS1) (Blázquez et al., 1998). Subsequent dephosphorylation of Tre6P by trehalose-6-phosphate phosphatase (TPP) produces trehalose. Despite its requirement for embryonic and vegetative development in plants (Eastmond et al., 2002), a detailed characterization of the TPS1 protein is still missing. In this issue of The Plant Cell, Fichtner et al. (2020) dissect the functional domains and localization of Arabidopsis TPS1 and assess their impact on metabolism.
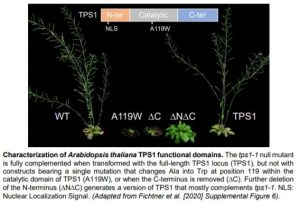 TPS1 has a central glucosyltransferase domain flanked by a putative N-terminal auto-inhibitory domain and a C-terminal TPP-like domain (see Figure). To dissect the contribution of each domain to TPS1 function, the authors tested various forms of TPS1 for complementation of the embryo-lethal tps1-1 mutant. They observed that catalytically dead TPS1 was not able to rescue tps1-1, indicating that enzymatic activity was essential for embryogenesis. However, restoring Tre6P synthesis by expressing E. coli TPS was sufficient to rescue tps1-1. By contrast, a single point mutation (A119W) resulted in dwarfed plants that never flowered, demonstrating that the catalytic domain also plays a function during vegetative growth (see Figure). Removing the N-terminal domain resulted in close to full complementation suggesting that this domain is dispensable for TPS1 function. On the contrary, TPS1 lacking its C-terminus (∆C) could not fully complement tps1-1. Interestingly, the additional removal of the N-terminus (∆N∆C) dampened this strong developmental phenotype, illustrating a functional interdependency between these domains.
TPS1 has a central glucosyltransferase domain flanked by a putative N-terminal auto-inhibitory domain and a C-terminal TPP-like domain (see Figure). To dissect the contribution of each domain to TPS1 function, the authors tested various forms of TPS1 for complementation of the embryo-lethal tps1-1 mutant. They observed that catalytically dead TPS1 was not able to rescue tps1-1, indicating that enzymatic activity was essential for embryogenesis. However, restoring Tre6P synthesis by expressing E. coli TPS was sufficient to rescue tps1-1. By contrast, a single point mutation (A119W) resulted in dwarfed plants that never flowered, demonstrating that the catalytic domain also plays a function during vegetative growth (see Figure). Removing the N-terminal domain resulted in close to full complementation suggesting that this domain is dispensable for TPS1 function. On the contrary, TPS1 lacking its C-terminus (∆C) could not fully complement tps1-1. Interestingly, the additional removal of the N-terminus (∆N∆C) dampened this strong developmental phenotype, illustrating a functional interdependency between these domains.
To investigate the expression pattern and intracellular localization of TPS1, Fichtner and colleagues generated transgenic lines expressing translational GUS and GFP fusions. They found that TPS1 is predominantly detected in the vasculature, the shoot apical meristem, and the phloem tissue. These observations agree with the importance of Tre6P in regulating sucrose levels and transport. Confocal microscopy showed that TPS1 is a nuclear protein containing an N-terminal nuclear localization signal (NLS). Consistently, removing the N-terminus of TPS1 resulted in a diffuse cytosolic localization. It is worth noting that this form of TPS1 still rescued tps1-1, suggesting that synthesis of Tre6P in the cytosol does not impair its metabolic and signaling functions.
With viable transgenic lines expressing different mutated versions of TPS1 in hand, the authors investigated their effects on sugar, nitrogen, and amino acid metabolism. Using mass spectrometry to analyze metabolites, they notably found that altering Tre6P levels led to changes in sucrose quantities, especially in the dwarfed plant lines (A119W and ∆C). These plants also accumulated two unknown disaccharide-phosphates, a potential pitfall of TPS1 catalytic infidelity. Finally, starved seedlings exposed to exogenous sucrose tended to over-accumulate sucrose when their Tre6P synthesis was partially compromised. Altogether, these metabolic analyses nicely illustrate the fine-tuning between Tre6P and sucrose levels, and their integration into the complex plant metabolism. In the future, it will be interesting to further dissect the metabolic nexus surrounding Tre6P and to understand how Tre6P is able to simultaneously be a key metabolite and act as a signaling molecule.
Sylvain Bischof
Department of Plant and Microbial Biology
University of Zürich, Switzerland
ORCHID ID: 0000-0003-2910-5132
REFERENCES
Lunn, J.E. et al. (2014). Trehalose metabolism in plants. Plant J. 79: 544-567.
Blázquez, M.A. et al. (1998). Isolation and molecular characterization of the Arabidopsis TPS1 gene, encoding trehalose-6-phosphate synthase. Plant J. 13: 685-689.
Eastmond, P.J. et al. (2002). Trehalose-6-phosphate synthase 1, which catalyses the first step in trehalose synthesis, is essential for Arabidopsis embryo maturation. Plant J. 29: 225-235.
Fichtner, F. et al. (2020). Functional features of TREHALOSE-6-PHOSPHATE SYNTHASE1 – an essential enzyme in Arabidopsis thaliana. Plant Cell DOI: https://doi.org/10.1105/tpc.19.00837.