A Molecular Gatekeeper of Algal Biofuel Synthesis
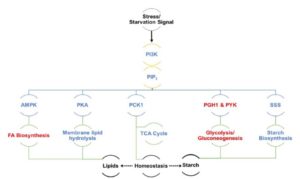 Algae undergo a complete metabolic transformation under stress by arresting cell growth, inducing autophagy, and hyper-accumulating biofuel precursors such as triacylglycerols and starch. However, the regulatory mechanisms behind this stress-induced transformation are still unclear. Understanding the signaling mechanism behind growth, starvation, and starvation-related responses is critical to the successful development of algal biofuels. Phosphatidylinositol 3-kinase (PI3K) signaling in higher eukaryotes has been connected to starvation-induced processes, including autophagy, lipid metabolism, and cell survival. Class III PI3K enzyme is the oldest class of this enzyme present in all known eukaryotes and is the only class in algae, yeasts, and plants. Ramanan et al. (10.1104/pp.17.01780) show that PI3K regulates the homeostasis of membrane lipids, triacylglycerols, starch, free fatty acids, and ATP in Chlamydomonas. Although PI3K is known to play a major role in autophagosome formation in higher eukaryotes, a PI3K knockdown Chlamydomonas mutant line reveals autophagy induction under stress, thereby ruling out any role of selective autophagic processes in the observed phenotype. From a biofuel perspective, this study sheds light on the regulatory mechanisms behind the tradeoff between growth and the accumulation of biofuel precursors. The manipulation of signal transduction pathways such as that governed by PI3K might provide solutions to several outstanding research bottlenecks in developing commercially viable algae-based biofuels.
Algae undergo a complete metabolic transformation under stress by arresting cell growth, inducing autophagy, and hyper-accumulating biofuel precursors such as triacylglycerols and starch. However, the regulatory mechanisms behind this stress-induced transformation are still unclear. Understanding the signaling mechanism behind growth, starvation, and starvation-related responses is critical to the successful development of algal biofuels. Phosphatidylinositol 3-kinase (PI3K) signaling in higher eukaryotes has been connected to starvation-induced processes, including autophagy, lipid metabolism, and cell survival. Class III PI3K enzyme is the oldest class of this enzyme present in all known eukaryotes and is the only class in algae, yeasts, and plants. Ramanan et al. (10.1104/pp.17.01780) show that PI3K regulates the homeostasis of membrane lipids, triacylglycerols, starch, free fatty acids, and ATP in Chlamydomonas. Although PI3K is known to play a major role in autophagosome formation in higher eukaryotes, a PI3K knockdown Chlamydomonas mutant line reveals autophagy induction under stress, thereby ruling out any role of selective autophagic processes in the observed phenotype. From a biofuel perspective, this study sheds light on the regulatory mechanisms behind the tradeoff between growth and the accumulation of biofuel precursors. The manipulation of signal transduction pathways such as that governed by PI3K might provide solutions to several outstanding research bottlenecks in developing commercially viable algae-based biofuels.