Transcriptional Switching Makes New Messages
Fukudome et al. explore the significance of Pol II C-terminal phosphorylation https://doi.org/10.1105/tpc.17.00331
By Akihito Fukudome and Hisashi Koiwa
Background: In animals and plants, gene expression begins with an enzyme called RNA polymerase II (Pol II), which produces ribonucleic acid (RNA) from the corresponding DNA sequence. Some, namely protein-coding messager RNAs, encode protein information; others are functional by themselves, therefore called non-coding RNAs. Pol II has a unique structure on its tail, the carboxyl-terminal domain (CTD), where a suite of reversible phosphorylation modifications orchestrates Pol II activity and the maturation process of newly synthesized RNAs. Previously, we found that the enzyme CTD-PHOSPHATASE LIKE 4 (CPL4) is responsible for removing phosphorylation from Pol II CTD in Arabidopsis. A transgenic Arabidopsis line deficient in CPL4 showed an abnormal protein-coding gene expression pattern, but the effects on non-coding RNA were unknown.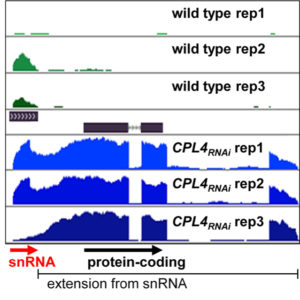
Question: We wanted to know how Pol II-CTD phosphorylation and CPL4 regulate protein-coding and non-coding gene expression in plants. Would environmental or developmental cues trigger changes in CTD phosphorylation and alter protein-coding or non-coding gene expression behavior?
Findings: In transgenic Arabidopsis plants deficient in CPL4 (CPL4RNAi), we found that Pol II does not stop properly when reading genes for a major class of non-coding RNA, namely small nuclear RNA (snRNA). When a protein-coding gene is present after a snRNA gene, the non-stop Pol II activity gave rise to a fusion RNA with protein-coding potential. Public database searches further identified multiple occasions of such snRNA-protein-coding RNA fusions in many plant species. Intriguingly, we also found salt stress can alter Pol II-CTD phosphorylation status and trigger this switching from non-coding RNA to protein-coding RNA in wild type Arabidopsis plants. An upstream snRNA promoter sequence could be an evolutionarily acquired regulatory mechanism for downstream protein-coding gene expression, as transposons contain the sequence element.
Next steps: An important next step will be to elucidate how such fusion events impact the biological function of downstream genes. Would it simply increase the expression level of the downstream gene, or change stability or mobility of the product RNAs? Furthermore, would it contribute to plant’s adaptation to stressful environment?
Fukudome, A., Sun, D., Zhang, X., and Koiwa, H. (2017). Salt Stress and CTD PHOSPHATASE-LIKE 4 Mediate the Switch Between Production of Small Nuclear RNAs and mRNAs. Plant Cell https://doi.org/10.1105/tpc.17.00331.