Tomato Genome Goes Nano
Schmidt et al. demonstrate that nanopore technology can be applied to plant genomes https://doi.org/10.1105/tpc.17.00521
By Schmidt, M. H.-W., Vogel, A., Denton, A. K., Bolger, A. M., Bolger, M. E., and Usadel, B.
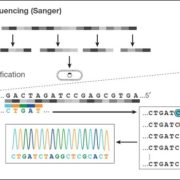
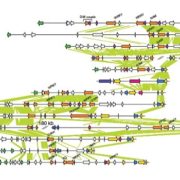
Background: An organism’s genome contains all the necessary information for its existence. Every genome consists of chromosomes, made of long DNA molecules built from four bases: adenine, guanine, cytosine and thymine. The genome of the wild tomato Solanum pennellii consists of 12 chromosomes totalling more than 1 billion bases. Oxford Nanopore Technology translates the changes in ion flow caused by the different bases in DNA fragments threaded through tiny pores about one nanometer in diameter directly into the corresponding DNA sequence in almost real time. To read the sequence of bases in the genome, we broke the chromosomes into pieces of several thousand bases, and these fragments were “read” using nanopore technology. Finally, a program finds overlaps in the different sequences and connects them to recreate the full chromosomal sequences.
Question: We wanted to test if complex genomes like that of a plant could be sequenced using nanopore technology. Before this, only less complicated genomes, mainly from fungi or bacteria, had been sequenced with nanopores.
Finding: We found that it is possible to sequence complex plant genomes using nanopores. Furthermore, nanopore sequencing makes it possible to do this relatively cheaply (about $25,000) with a relatively small working group (just 3 people in the lab) in relatively short time (about 8 months). Such a project typically would have taken several years and involved many large working groups. Unlike other sequencing technologies, nanopore sequencing furthermore does not require a big capital investment for the sequencing device itself, as the Nanopore sequencer costs only about $1000 in comparison to other sequencers that often cost more than $100,000.
Next steps: We are working on increasing the lengths of the DNA fragments that are sequenced from about 12 kilobases bases to 30 or even 50 kilobases. Longer fragments would simplify reading the genome and will enable reading genomes of plants which are even larger than that of the tomato genome.
Schmidt, et al. (2017). De novo assembly of a new Solanum pennellii accession using nanopore sequencing. Plant Cell DOI: https://doi.org/10.1105/tpc.17.00521