Thrown for a Loop: How RNase H1 and DNA Gyrases Limit R-loops and Maintain Genome Stability in Chloroplasts
We all know that DNA is the stable nucleic acid, in comparison to its flighty, unstable cousin RNA, right? Well, unusual things happen when metabolic processes require DNA to unwind from its stable, redundant double-helical form. For example, during transcription, the RNA that exits RNA polymerase can pair with the template DNA strand, leaving the non-templated strand as single-stranded DNA (ssDNA), forming a structure called an R-loop (reviewed in Gaillard and Aguilera, 2016). R-loops function in chromatin structure and in regulating transcription; for example, an R-loop regulates the antisense long, non-coding RNA COOLAIR that transcribes from FLOWERING LOCUS C (Sun et al. 2013). Conversely, R-loops can also affect the stability of the nuclear or organellar genomes. For example, the loop of ssDNA may be vulnerable to mutation or breakage; R-loops can also prevent or detain the progress of the DNA replication fork.
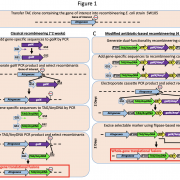
Cells and organelles have multiple mechanisms to mitigate the detrimental effects of R-loop formation. For example, RNase H enzymes can degrade the RNA part of the RNA–DNA hybrid, thereby allowing the ssDNA to pair with its complementary DNA strand and resolving the R-loop. To explore the functions of RNase H enzymes in plants, Yang et al. (2017) comprehensively identified RNase H1 enzymes encoded by the Arabidopsis thaliana genome. Phylogenetic analysis and examination of fluorescent protein fusions showed that higher plants have evolved three RNase H1-like proteins localized to the subcellular compartments that contain DNA (nucleus, mitochondria, and chloroplasts).
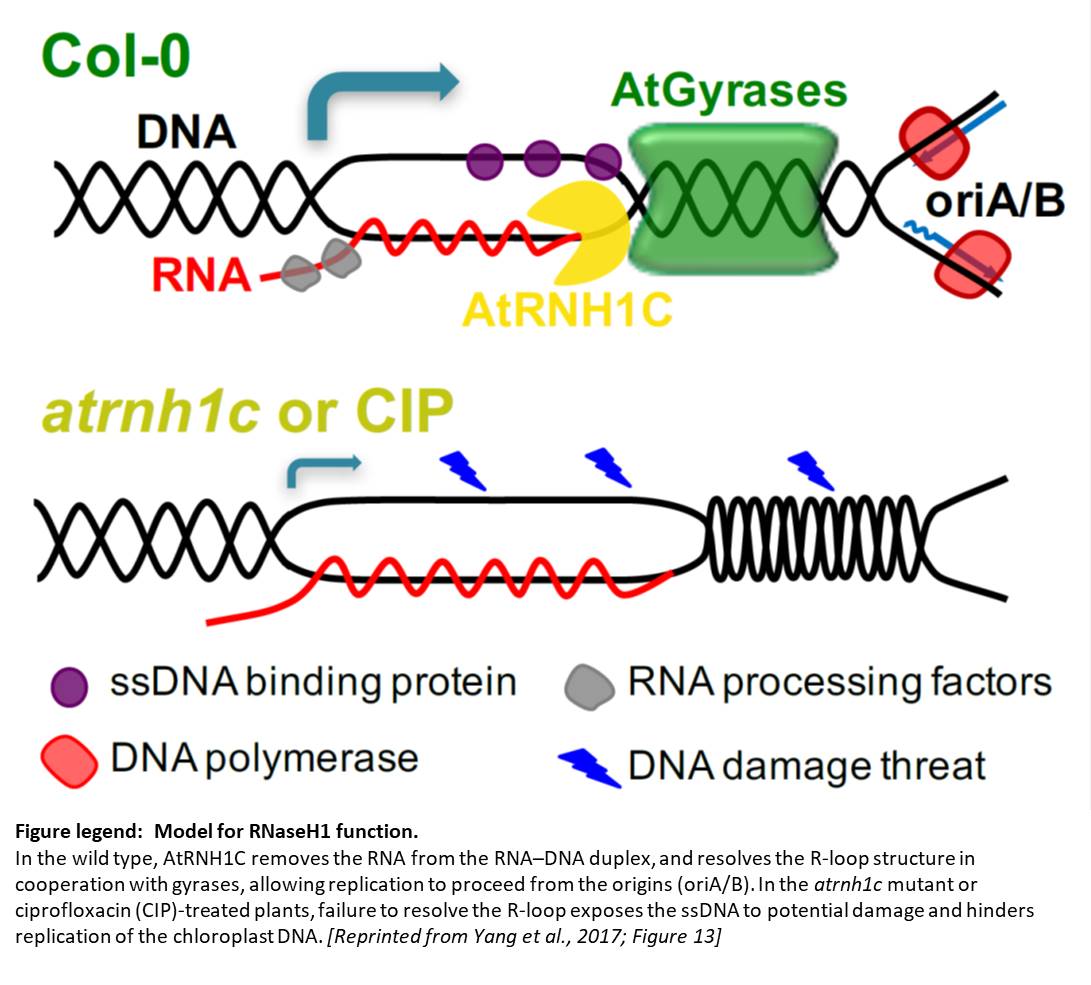
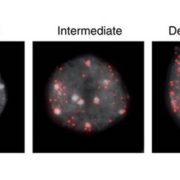
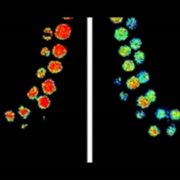
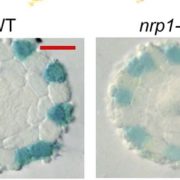
Despite being located in close proximity to the photosynthetic apparatus (which, in moving high-energy electrons can also generate free radicals), and despite being heavily transcribed, the chloroplast genome remains stable, even compared to the nuclear genome. To examine the role of RNase H1 in the chloroplast, the authors focus on the chloroplast-localized form, AtRNH1C. Knockout atrnh1c mutants (T-DNA insertion and CRISPR/Cas9 mediated) showed severe chloroplast abnormalities and a pale-green phenotype. In vitro activity assays showed that AtRNH1C has RNase H activity, being able to degrade the RNA strand of an RNA–DNA hybrid. Moreover, examination of the chloroplasts in the atrnh1c mutants by DNA–RNA immunoprecipitation, digestion-sensitive R-loop profiling, and R-loop footprinting showed that the chloroplast DNA contained much higher levels of R-loops and more extensive R-loops, and a substantial increase in degraded and rearranged chloroplast DNA in the atrnh1c mutants, compared with wild-type plants.
Affinity purification of tagged AtRNH1C and mass spectrometry showed that AtRNH1C interacts with multiple proteins involved in genome structure and stability, including an organelle-localized Arabidopsis DNA gyrase (a type II topoisomerase that introduces negative supercoiling). Further examining the interaction of AtRNH1C with DNA gyrase, the authors found that nuclease treatment disrupted the interaction of these proteins, indicating the requirement for DNA or RNA in the AtRNH1C–gyrase interaction. Also, treatment with the gyrase inhibitors ciprofloxacin or novobiocin produced an additive effect on the atrnh1c mutants, indicating that AtRNH1C and DNA gyrase may collaborate to resolve R-loops.
The authors used two-dimensional gel electrophoresis to examine the effect of AtRNH1C and DNA gyrase on the replication of DNA from the conserved origins of replication in the inverted repeats of the chloroplast DNA. Indeed, they found fewer replication intermediates in the atrnh1c mutants and ciprofloxacin-treated cells, indicating a blockage of replication. They also observed reduced transcription of the ribosomal DNA. Chromatin immunoprecipitation showed that AtRNH1C physically associates with these inverted repeat regions and that association decreased in plants treated with gyrase inhibitors. Therefore, these two enzymes have key functions for both genome stability and ribosomal DNA transcription.
Intriguingly, ciprofloxacin can induce DNA double-strand breaks and the additive effect of ciprofloxacin in the atrnh1c mutants indicates that AtRNH1C may function in repair of the chloroplast DNA, as emerging research indicates that RNA–DNA hybrids may participate in DNA repair. Examination of how AtRNH1C interacts with its various partners, and dissecting its various functions will prove an intriguing prospect for future research. Moreover, determining how RNase H and its partners interfere with “bad” R-loops while not interfering with the important regulatory functions of R-loops remains an outstanding question.
REFERENCES
Gaillard, H., and Aguilera A. (2016) Transcription as a Threat to Genome Integrity. Annual Review of Biochemistry 85:291–317 https://doi.org/10.1146/annurev-biochem-060815-014908
Sun, Q., Csorba, T., Skourti-Stathaki, K., Proudfoot, N.J., Dean, C. (2013) R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 340(6132):619-621 doi: 10.1126/science.1234848
Yang, Z., Hou, Q., Cheng, L., Xu, W., Hong, Y., Li, S., and Sun, Q. (2017) RNase H1 Cooperates with DNA Gyrases to Restrict R-loops and Maintain Genome Integrity in Arabidopsis Chloroplasts. Plant Cell 10.1105/tpc.17.00305.