A Histone Chaperone and a Specific Transcription Factor Modulate GLABRA2 Expression in Root Hair Development
IN BRIEF by Jennifer Mach [email protected]
 To navigate its essential function of producing mRNAs, RNA polymerase II (Pol II) must navigate the thread of DNA, which winds around thousands of nucleosomes. If you’ve ever tried to use a sewing machine but got your bobbin thread tangled, then the task faced by Pol II may seem impossible. However, Pol II has help: Histone chaperones and chromatin-remodeling complexes remove nucleosomes from its path; they also slide nucleosomes, prevent the aggregation of free histones with DNA, and exchange nucleosomes with histone variants (reviewed in Venkatesh and Workman, 2015). Some chaperones interact preferentially with different histones; for example, H2A/H2B chaperone interact with the H2A/H2B dimers that flank the (H3-H4)2 tetramer in the histone core.
To navigate its essential function of producing mRNAs, RNA polymerase II (Pol II) must navigate the thread of DNA, which winds around thousands of nucleosomes. If you’ve ever tried to use a sewing machine but got your bobbin thread tangled, then the task faced by Pol II may seem impossible. However, Pol II has help: Histone chaperones and chromatin-remodeling complexes remove nucleosomes from its path; they also slide nucleosomes, prevent the aggregation of free histones with DNA, and exchange nucleosomes with histone variants (reviewed in Venkatesh and Workman, 2015). Some chaperones interact preferentially with different histones; for example, H2A/H2B chaperone interact with the H2A/H2B dimers that flank the (H3-H4)2 tetramer in the histone core.
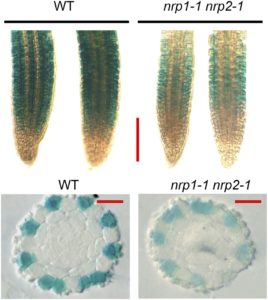
Histone chaperones affect transcription, DNA replication, and DNA repair and play important roles in plant growth and development (reviewed in Zhou et al., 2015). For example, mutants of the histone H2A/H2B chaperone genes NUCLEOSOME ASSEMBLY PROTEIN1-RELATED PROTEIN1 (NRP1) and NRP2 produce ectopic root hairs. Root hairs develop from single epidermal cells (H-cells). In Arabidopsis thaliana, H-cells develops over the junctions between pairs of cortical cells; epidermal cells not directly over the junction (N-cells) do not generally form root hairs. The homeodomain-leucine zipper transcription factor GLABRA2 (GL2) suppresses formation of root hairs in N-cells, and root hair specification involves changes in GL2 transcription. Zhu et al. (2017) examined the role of NRP1 and NRP2 in root hair development and found that the nrp1-1 nrp2-1 mutants have lower GL2 expression, although GL expression occurs in the same cells.
The basic helix-loop-helix transcription factor WEREWOLF (WER) forms part of a complex that activates GL2 expression in N-cells, and the transcript levels of WER and other upstream regulators of GL2 remained unchanged in the nrp1-1 nrp2-1 mutants. Also, the wer-1 nrp1-1 nrp2-1 triple mutants had more root hairs than the nrp1-1 nrp2-1 double mutants, similar to the wer-1 single mutants, suggesting that wer-1 is epistatic. The authors next used chromatin immunoprecipitation-PCR to show that association of NRP1 with the GL2 promoter decreased significantly in the wer-1 mutants. Pull-down and bimolecular fluorescence complementation assays showed that WER and NRP1 directly interact; size-exclusion chromatography indicated that WER and NRP1 form a 1:1 complex and that NRP1 forms dimers. Indeed, determination of the crystal structure of NRP1 showed that NRP1 forms dimers through an N-terminal α-helix. Mutations that disrupt the ability of NRP1 to form dimers and mutants that disrupt its acidic C terminus decrease NPR1’s interaction with histones and with WER1. Moreover, these mutants do not rescue the nrp1-1 nrp2-1 phenotype.
These results linked the NRP1 and NRP2 histone chaperones with WER; the authors next explored how these factors affect chromatin structure at GL2. They found that the GL2 promoter had higher histone occupancy and nucleosome density (and was less accessible to micrococcal nuclease) in the nrp1-1 nrp2-1 mutants than in the wild type. Other loci, such as ACTIN2 and FLOWERING LOCUS C, did not show a difference between the mutant and wild type. Finally, electrophoretic mobility shift assays showed that the presence of histones decreased binding of WER to its target, but NRP1 removed the histones and allowed WER to bind.
This study used a wide-range of techniques, including genetics, molecular biology, and crystallography, to examine how the interaction of histone chaperones with the WER transcription factor targets these chaperones to GL2 and alters the chromatin there. Determining how generally this mechanism occurs, what other chromatin factors affect GL2, and how NRPs regulate other loci will provide intriguing topics for future work.
Venkatesh, S., Workman, J.L. (2015). Histone exchange, chromatin structure, and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 16: 178–189.
Zhou, W., Zhu, Y., Dong, A., Shen, W.H. (2015). Histone H2A/H2B chaperones: from molecules to chromatin-based functions in plant growth and development. Plant J. 83: 78–95.
Zhu, Y., Rong, L., Luo, Q., Wang, B., Zhou, N., Yang, Y., Zhang, C., Feng, H., Zheng, L., Shen, W.-H., Ma, J., Dong, A. (2017). The histone chaperone NRP1 interacts with WEREWOLF to activate GLABRA2 in Arabidopsis root hair development. Plant Cell 29: 260–276.




Leave a Reply
Want to join the discussion?Feel free to contribute!