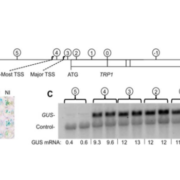
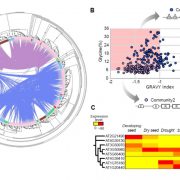
The evolution of CHROMOMETHYLASES and gene body DNA methylation in plants
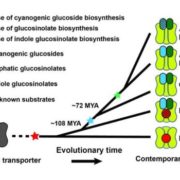
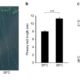
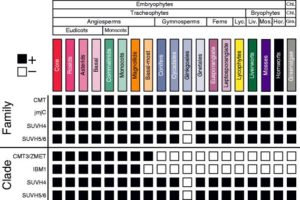 In land plant genomes, transposable elements (TEs) are ubiquitously methylated in CG and non-CG sequence contexts. Apart from methylation of TEs, DNA methylation also occurs on bodies of actively transcribed genes, typically in the CG context, with very low or no non-CG methylation, and is referred to as gene body methylation (gbM). To begin to understand the evolution, origins, and functions of gbM and its relationship to CHROMOMETHYLTRANSFERASE (CMT) proteins, Bewick et al., used data from more than 1000 transcriptomes and 77 methylomes for phylogenetic and methylation variation analysis. Their extensive taxonomic sampling redefined the CMT clade and showed that CMT genes originated prior to the evolution of land plants. CMT3 has been shown to maintain methylation at CHG sites, and CMT2 methylates CHH sites in the heterochromatin, while CMT1 is non-functional. Apart from maintaining non-CG methylation at TEs in the pericentromeric regions, CMTs are also important for CG methylation within gbM genes. In Brassicaceae, particularly in the clade containing B. oleracea, B. rapa, and S. parvula, CMT3 has a higher rate of molecular evolution measured by dN/dS compared to other eudicots, potentially disrupting CMT3 function to varying degrees. The authors also show the tightly correlated evolution of CMT3 and IBM1, a histone demethylase which preferentially demethylates at H3K9, suggesting their involvement in the gbM pathway. These results open avenues for future work to help better understand the consequences of and the relationship between DNA gene body methylation and gene expression. Genome Biol. 10.1186/s13059-017-1195-1 (Contributed by Sunil Kumar K R)
In land plant genomes, transposable elements (TEs) are ubiquitously methylated in CG and non-CG sequence contexts. Apart from methylation of TEs, DNA methylation also occurs on bodies of actively transcribed genes, typically in the CG context, with very low or no non-CG methylation, and is referred to as gene body methylation (gbM). To begin to understand the evolution, origins, and functions of gbM and its relationship to CHROMOMETHYLTRANSFERASE (CMT) proteins, Bewick et al., used data from more than 1000 transcriptomes and 77 methylomes for phylogenetic and methylation variation analysis. Their extensive taxonomic sampling redefined the CMT clade and showed that CMT genes originated prior to the evolution of land plants. CMT3 has been shown to maintain methylation at CHG sites, and CMT2 methylates CHH sites in the heterochromatin, while CMT1 is non-functional. Apart from maintaining non-CG methylation at TEs in the pericentromeric regions, CMTs are also important for CG methylation within gbM genes. In Brassicaceae, particularly in the clade containing B. oleracea, B. rapa, and S. parvula, CMT3 has a higher rate of molecular evolution measured by dN/dS compared to other eudicots, potentially disrupting CMT3 function to varying degrees. The authors also show the tightly correlated evolution of CMT3 and IBM1, a histone demethylase which preferentially demethylates at H3K9, suggesting their involvement in the gbM pathway. These results open avenues for future work to help better understand the consequences of and the relationship between DNA gene body methylation and gene expression. Genome Biol. 10.1186/s13059-017-1195-1 (Contributed by Sunil Kumar K R)