The Battle Between Modifications on Chromatin
Dong, LeBlanc, Poulet et al. investigate genomic stability in Arabidopsis. The Plant Cell (2020) https://bit.ly/3nhgdQ5
By Jie Dong, Chantal LeBlanc, Axel Poulet, and Yannick Jacob
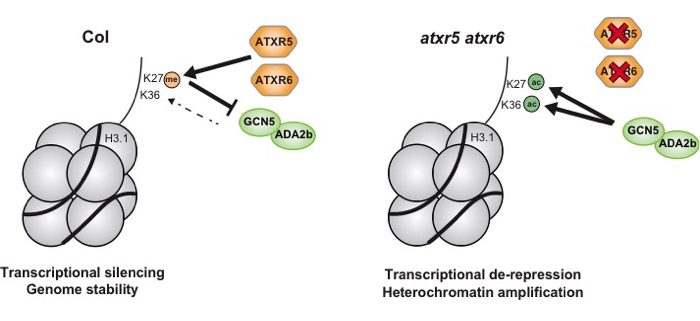
Background: DNA is the genetic material used by all living organisms on Earth. The faithful replication of DNA ensures the precise transfer of genetic information from mother cell to daughter cells as well as from parents to progeny. In eukaryotes, DNA is compacted into chromatin, which consists of DNA and the histone proteins H1, H2A, H2B, H3, and H4. These histone proteins can be modified with specific chemical groups, often referred to as epigenetic marks, which affect cellular processes occurring at the DNA level (e.g. transcription). Two common epigenetic modifications on histones are methylation (me) and acetylation (ac) on lysine residues. Is it known that the absence of one methyl group (me1) on lysine (K) 27 of histone H3 (H3K27me1), normally deposited by the methyltransferases ATXR5 and ATXR6 (ATXR5/6), causes transcriptional mis-regulation of some transposons and genomic instability in plants.
Question: We wanted to know what other modifications besides H3K27me1 are involved in regulating genomic stability in plants, and which enzymes catalyze them.
Findings: We found that H3K27ac and H3K36ac, which are deposited by the histone acetyltransferase GCN5 and its partner ADA2b, contribute to genomic instability in the absence of H3K27me1 in the model plant Arabidopsis thaliana. When we removed GCN5 or ADA2b from a plant already lacking ATXR5/6, the phenotypes associated with the loss of H3K27me1 were almost completely suppressed. In addition, we found that in the absence of H3K27me1, H3K27ac and H3K36ac would accumulate at the regions of the genome where genomic instability occurred. Finally, we discovered that the presence of K27me1 on histone H3 directly prevents the addition of acetyl groups to K27 and K36 by GCN5-ADA2b.
Next steps: To better understand the underlying mechanisms responsible for the maintenance of genomic stability, scientists will have to identify the downstream proteins that are interacting with modified (e.g. methylated or acetylated) and unmodified H3K27 and H3K36 and clarify the relationship between all the phenotypes associated with the loss of H3K27me1.
Jie Dong, Chantal LeBlanc, Axel Poulet, Benoit Mermaz, Gonzalo Villarino, Kimberly M. Webb, Valentin Joly, Josefina Mendez, Philipp Voigt and Yannick Jacob. (2021). H3.1K27me1 maintains transcriptional silencing and genome stability by preventing GCN5-mediated histone acetylation. Plant Cell https://bit.ly/3nhgdQ5