Highly Diverse Plant Immune Receptors Form a Species-wide Reservoir of New Recognition Specificities
By Daniil M. Prigozhin1 and Ksenia V. Krasileva2
1Lawrence Berkeley National Laboratory
2University of California Berkeley
Background: Harmful bacteria, fungi, oomycetes, insects, nematodes, and viruses threaten plant health and can devastate agricultural crops. Plants have an innate immune system that protects them from disease. Unlike human immune proteins, which can rapidly generate new recognition specificities in response to infection or immunization, plant immune proteins do not change over the lifetime of a single organism. The generation of plant immune receptor diversity must therefore occur at the population level. How plants learn to recognize newly emergent threats remains to be fully explored.
Question: We wanted to find out where the new recognition specificities in plant immunity come from.
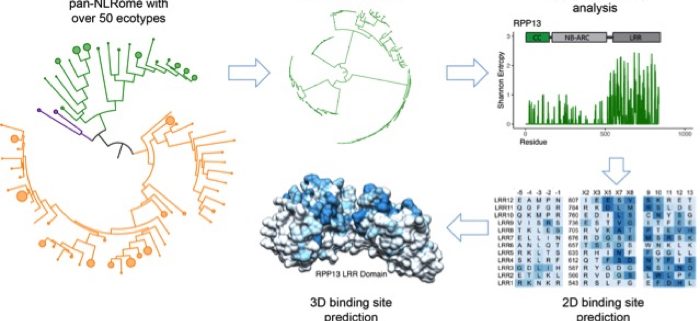
Findings: In this study, we used data already available for the model dicot and model monocot plants Arabidopsis and Brachypodium, respectively. In each species, we compared immune receptors across over 50 divergent plant varieties. We developed new methods to analyze plant immune receptors that rely on careful reconstruction of their natural history and on using protein alignment entropy to find variable positions. We observed that some immune protein families contain receptors that vary little from ecotype to ecotype while others are highly variable. In the families with high sequence diversity, the observed differences clustered together on the protein surfaces, allowing us to predict the regions used to bind to the pathogen targets in each receptor. We concluded that at the population level, the highly variable receptor families form a reservoir of new specificities, while at least some of the invariable receptors serve as a repository of the successful variants derived over many years of evolution.
Next steps: These findings will help us look for natural forces that determine the patterns of evolution of different immune receptor families. They will also guide future efforts to engineer disease resistance in crop species in order to improve plant health.
Daniil M Prigozhin and Ksenia V Krasileva (2021). Analysis of intraspecies diversity reveals a subset of highly variable plant immune receptors and predicts their binding sites. Plant Cell https://academic.oup.com/plcell/advance-article/doi/10.1093/plcell/koab013/6119334