Targeted A-to-G base editing of chloroplast DNA in plants
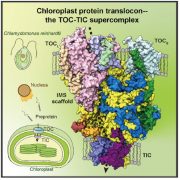
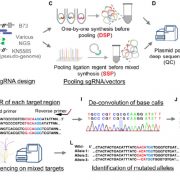
 The chloroplast has its own genome which encodes proteins needed for chloroplast function. Editing these genes via CRISPR-Cas9 is difficult due to challenges in targeting guide RNAs to the chloroplast. Instead, base editors can be used, but these were originally developed for cytosine to thymine edits in mitochondria. Here, Mok et al. have used similar ideas to develop a chloroplast targeted adenine to guanine (A to G) base editor. Their editor consists of pairs of chloroplast targeted TALE (transcription activator-like effector) DNA-binding arrays linked to deaminases. When designed to target the photosynthetic gene psaA in Arabidopsis, there was specific editing of adenine to guanine with up to 99% efficiency, and this was accompanied by the expected albino phenotype. Interestingly, one plant was mosaic-like, with both green and pale leaves. The pale leaves had edits at the psaA start codon in 95% of cases, whereas in the green leaves the editing efficiency was 0.1%, demonstrating that the pale phenotype is in fact caused by the editing. Remarkably, these edits persisted in the subsequent generation and in some cases without the presence of the transgene. Therefore, the specific heritable edits generated by this base editor will make it easier to alter the chloroplast genome to study gene function. (Summary by Rose McNelly @Rose_McN) Nature Plants 10.1038/s41477-022-01279-8
The chloroplast has its own genome which encodes proteins needed for chloroplast function. Editing these genes via CRISPR-Cas9 is difficult due to challenges in targeting guide RNAs to the chloroplast. Instead, base editors can be used, but these were originally developed for cytosine to thymine edits in mitochondria. Here, Mok et al. have used similar ideas to develop a chloroplast targeted adenine to guanine (A to G) base editor. Their editor consists of pairs of chloroplast targeted TALE (transcription activator-like effector) DNA-binding arrays linked to deaminases. When designed to target the photosynthetic gene psaA in Arabidopsis, there was specific editing of adenine to guanine with up to 99% efficiency, and this was accompanied by the expected albino phenotype. Interestingly, one plant was mosaic-like, with both green and pale leaves. The pale leaves had edits at the psaA start codon in 95% of cases, whereas in the green leaves the editing efficiency was 0.1%, demonstrating that the pale phenotype is in fact caused by the editing. Remarkably, these edits persisted in the subsequent generation and in some cases without the presence of the transgene. Therefore, the specific heritable edits generated by this base editor will make it easier to alter the chloroplast genome to study gene function. (Summary by Rose McNelly @Rose_McN) Nature Plants 10.1038/s41477-022-01279-8