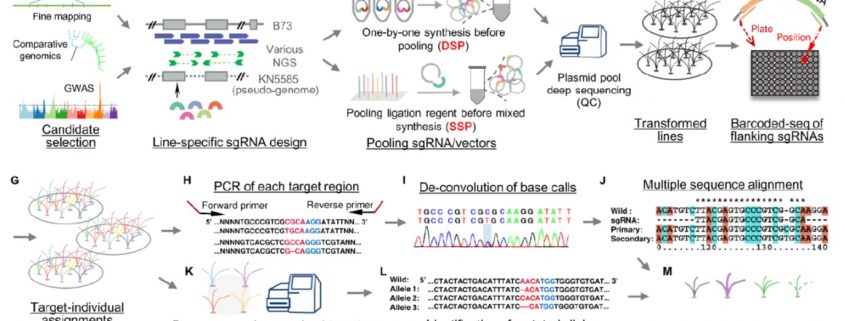
High-throughput CRISPR/Cas9 mutagenesis streamlines trait gene identification in maize (Plant Cell)
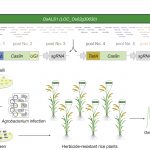
Maize has provided some fascinating mutants and developmental insights, but its genomic complexity has made it more difficult (for example as compared to rice) to identify agronomically important genes. Liu et al. describe a new high-throughput method to integrate forward- and reverse-genetics to identify genes in maize and other crops with complex genomes. They selected 1244 potentially interesting candidate genes that had been identified from either classical mapping or comparative genomics. They made six CRISPR libraries of sgRNAs, which they transformed into maize embryos using Agrobacterium, and sequenced the transformants using primers from the conserved plasmid backbone to identify induced sequence variation. Sixty percent of the changes were small deletions (1-65 bp), with the remainder being small insertions or base substitutions. They found only a small number of off-target mutations, but a high rate of mosaicism. The data produced from this work will help others design efficient mutagenesis approaches and anticipate the outcomes, and, the “library of edited genes provides an unprecedented resource for further detailed functional genomics.” (Summary by Mary Williams) Plant Cell 10.1105/tpc.19.0093
[altmetric doi=”/10.1105/tpc.19.00934″ details=”right” float=”right”]