Summer fun: how plants beat the heat
By Adam Phillips. Reprinted from It Ain’t Magic, The RIKEN Global Communications Team https://itaintmagic.riken.jp/hot-off-the-press/plants-beat-heat
It seems like I’ve been writing a lot about plants recently. The truth is that I hardly have enough time to write about all the cool plant studies going on. The study I’m writing about today is cool because it deals with the discovery of a gene related to how plants respond to extreme heat. The the Metabolomics Research Group at RIKEN CSRS have named the gene HIL1, and it is believed to function by preventing the destabilization of chloroplast membranes that occurs at very high temperatures.
We all know how uncomfortable it is to be stuck outside on a sweltering hot day. Now, imagine how bad it would be if you were a soybean or tomato plant without any chance of moving inside. Eventually your leaves might become bleached of color due to chloroplast membrane damage, and if you did not get any relief, you might die. Fortunately for plants, they do have a natural defense against this type of stress that involves modifying plant fats that make up chloroplast membranes. When heat causes chloroplast membranes to destabilize, polyunsaturated fatty acids are removed from the membrane lipids, which stabilizes the membranes. The team at RIKEN found the gene responsible for this process, and they did so rather quickly because of their innovative approach.
“We hypothesized that breaking down of chloroplastic lipids would be controlled by some unidentified genes that encode lipases localized in chloroplasts and which are induced by heat stress,” explains first author Yasuhiro Higashi. “We used several public omics databases to isolate candidate genes involved in heat-stress responses, predict where the genes are expressed in the plants, and to find gene homologues in several plants species.”
Lipases are proteins that break apart lipids. A transcriptome analysis identified candidate lipase genes, and proteomics databases were able to narrow down the search by predicting which genes were localized in chloroplasts. To test the most likely candidate gene—which they subsequently named Heat Inducible Lipase 1 (HIL1)—they created a hil1 mutant Arabidopsis plant in which the gene was disrupted, and compared mutant responses to heat stress with those of normal plants.
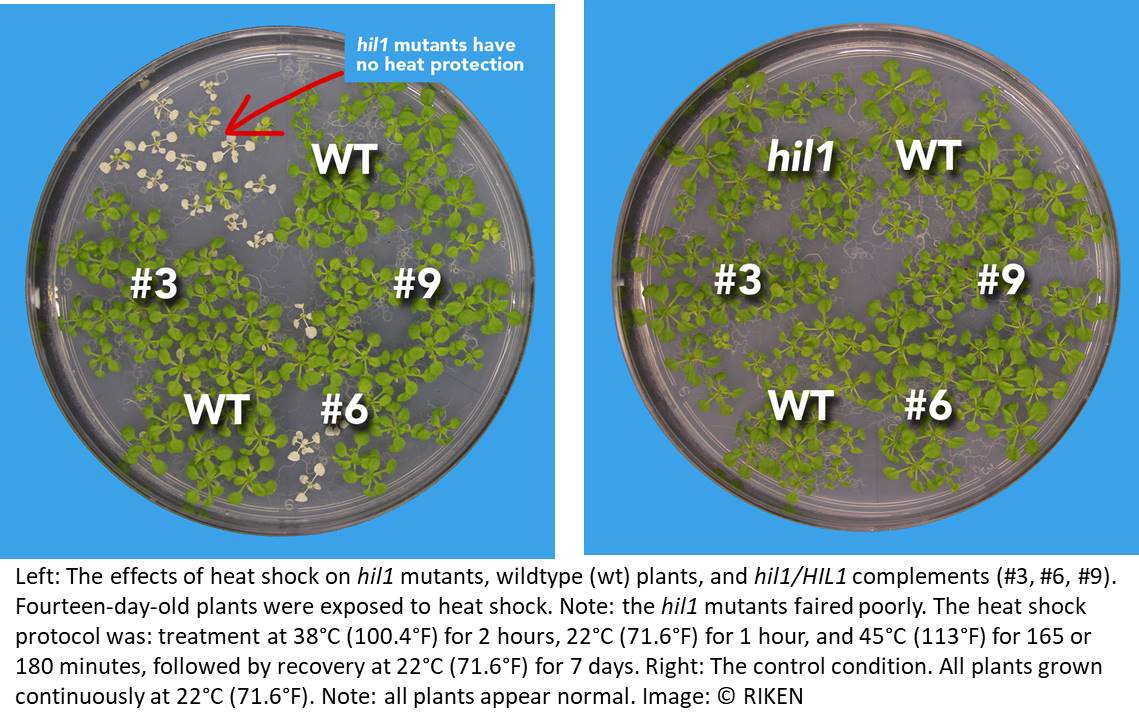
They found that the mutant plants were more sensitive to heat, meaning that their condition worsened more than control plants in extreme heat and their survival rate was much lower. Closer examination showed that membrane remodeling in the mutant chloroplasts was partially disrupted, and not all polyunsaturated fatty acids were removed from a key glycerolipid.
These experiments were performed in the plant Arabidopsis, the most common plant used in plant research. The team also did a gene coexpression analysis using public microarray data, which revealed a conserved network associated with HIL1 in several plant species, including rice, tomatoes, corn, and soybeans. “We are interested in discovering the function of the HIL1 homologous genes in major crop species,” notes Higashi. “We hope our knowledge about HIL1 might aid in the development of new heat stress-tolerant plant varieties, which could help improve crop yield in hot environments.”
Team leader Kazuki Saito is also looking at the big picture. “This natural response to heat is not new, but understanding how it works is a step forward in fighting global warming,” he says. “Our study certainly contributes to the Sustainable Development Goals (STGs) set by the UN in 2015, in particular, goals 2 and 13—Climate Action and Zero Hunger.” ✅
Further reading
Higashi et al. (2018) HEAT INDUCIBLE LIPASE1 remodels chloroplastic monogalactosyldiacylglycerol by liberating α-linolenic acid in Arabidopsis leaves under heat stress. Plant Cell. doi: 10.1105/tpc.18.00347.