Size no longer matters: Cloning algal genes just became easier
By Tom Z. Emrich-Mills, Gary Yates, and Luke C.M. Mackinder
Background: Chlamydomonas reinhardtii is a single-celled alga that shares similar cellular machinery to plants, making it a useful model for studying how plant cells function. Like all organisms, the DNA present in a Chlamydomonas cell encodes all the proteins required for the cell to survive and thrive. To discover the function of a particular protein, bioscientists often first copy the DNA that encodes the protein and insert it into a circular piece of DNA called a plasmid (known as cloning). Cloning can be difficult because PCR–the dominant cloning method–becomes inefficient if the DNA sequence in question is very long or repetitive. Unfortunately, a large proportion of Chlamydomonas proteins are encoded by long and complicated genes, hampering progress in understanding this organism.
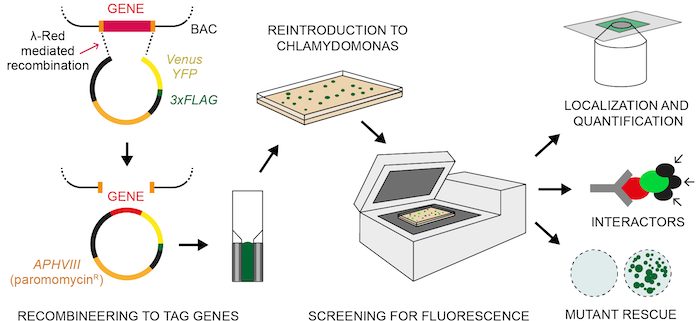
Question: We set out to discover whether an alternative method for cloning DNA called recombineering could be applied to Chlamydomonas genes for large-scale discovery of protein location and function. Recombineering is fundamentally different from PCR and does not share the same limitations caused by the DNA sequence.
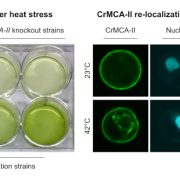
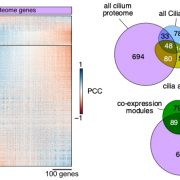
Findings: Our method successfully enabled the cloning of very large and repetitive Chlamydomonas genes. We also found that our method was scalable and could achieve a success rate of 77%, almost double that of previous PCR-based cloning, when applied to 203 gene targets. Targets were chosen due to their involvement in the uptake of CO2 into Chlamydomonas cells. The plasmids we designed contain the sequences for a range of fluorescent protein tags, which allows the encoded proteins of these newly cloned genes to be visualized under a microscope to reveal their cellular location. Our findings open the door to understanding the functions of hundreds of proteins that were previously problematic to work with due to their DNA sequences. Scientists working on many different aspects of algal biology will be able to apply or adapt our method to further their work.
Next steps: We aim to use the method to rapidly advance our understanding of how Chlamydomonas cells efficiently acquire CO2 from their environment.
Tom Z. Emrich-Mills, Gary Yates, James Barrett, Philipp Girr, Irina Grouneva, Chun Sing Lau, Charlotte E. Walker, Tsz Kam Kwok, John W. Davey, Matthew P. Johnson and Luke C.M. Mackinder. (2021). A recombineering pipeline to clone large and complex genes in Chlamydomonas. Plant Cell https://doi.org/10.1093/plcell/koab024