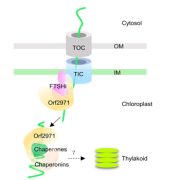
Systematic characterization of gene function in a photosynthetic organism (bioRxiv)
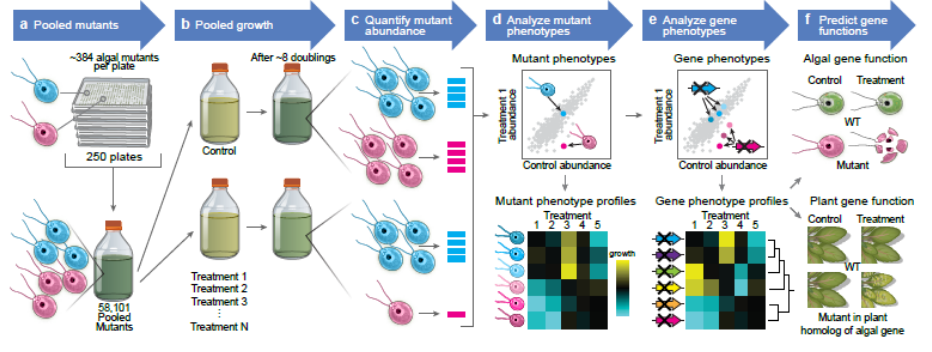
Vilarrasa-Blasi and coworkers described their impressive work on a huge barcoded Chlamydomonas collection, in which they screened almost 60,000 insertion mutants grown under a wide range of environmental and chemical stress conditions. The authors assessed genotype-phenotype specificity for almost 80% of C. reinhardtii genes, represented by one or multiple allelic variants, and shed new light into the function of uncharacterized genetic determinants, including conserved genes involved in photosynthesis and novel factors underpinning CO2-concentrating mechanism. The identification of mutants showing sensitivity to Latrunculin B (LatB), a defensive molecule that interferes with actin polymerization, represents a clear example of this powerful approach. In C. reinhardtii, F-actin homeostasis pathway relies on the activation of NOVEL ACTIN-LIKE PROTEIN1 (NAP1), which is resistant to LatB, mediated by LatB-SENSITIVE (LAT1-LAT3) factors. In this study, the authors identified three novel components (LAT5-7) and demonstrated the conservation of the pathway underlying cytoskeleton integrity in the model plant Arabidopsis thaliana. High-throughput data generated in green algae provide important tools that will pave the way for functional genomics in land plant species, especially for the characterization of unknown genes involved in essential processes and adaptation to different environments. (Summary and image adaptation by Michela Osnato @michela_osnato) bioRxiv 10.1101/2020.12.11.420950