Chlamydomonas genomics: no longer going it alone
By Rory J. Craig, Institute of Evolutionary Biology, University of Edinburgh
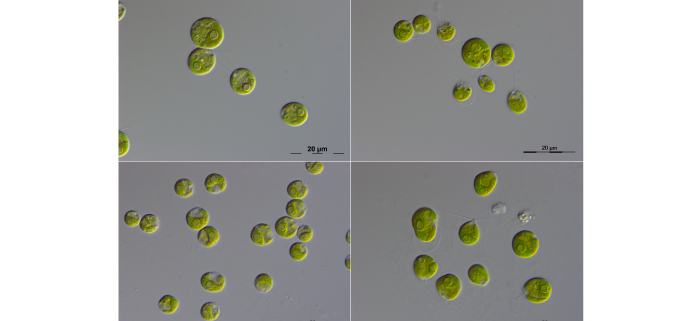
Background: The single-cell green alga Chlamydomonas reinhardtii is one of the most important species for the study of plant cell biology. However, unlike most land plants, there are currently no genome assemblies or gene annotations available for close relatives of C. reinhardtii. This situation limits comparative analyses at the protein level, for example between C. reinhardtii and its somewhat distant multicellular relative Volvox carteri, or to more distant relatives such as Arabidopsis thaliana or human. While protein-based approaches are fundamental to addressing many questions, several analyses can only be performed at the nucleotide level: characterizing general features of genomic architecture, improving gene annotations by identifying new genes based on sequence conservation, and identifying functional noncoding sequences, including regulatory elements.
Question: We aimed to address the lack of genomic resources for close relatives of C. reinhardtii by producing high-quality genome assemblies and gene annotations for two species in the genus Chlamydomonas: C. incerta and C. schloesseri. As these are its only known close relatives, we also sequenced and annotated a genome assembly for the more distant relative Edaphochlamys debaryana.
Findings: The genomes of C. incerta and C. schloesseri are highly co-linear with the C. reinhardtii genome, although they are slightly larger and more repetitive. We characterized the putative centromeric repeat in C. reinhardtii as a retrotransposon related to Zepp, the centromeric component of the very distantly related alga Coccomyxa subellipsoidea. C. incerta and C. schloesseri likely share centromeric locations and structures with C. reinhardtii, and Zepp-like elements may be associated with centromeres in other green algae. Using comparative genomics approaches, we improved C. reinhardtii gene annotations, identifying more than 100 new genes, several hundred existing genes that are likely false positives, and ~10 Mb of conserved noncoding sequence.
Next steps: With the resources presented in this study, we are taking only the first steps towards realizing the full power of comparative genomics for C. reinhardtii, and many of the analyses we performed should be substantially improved by adding more genomes, which is currently limited by the availability of samples. Efforts should thus be redoubled to discover new Chlamydomonas species.
Citation:
Rory J. Craig, Ahmed R. Hasan, Rob W. Ness & Peter D. Keightley (2021). Comparative genomics of Chlamydomonas. The Plant Cell, https://doi.org/10.1093/plcell/koab026