Single-cell RNA sequencing reveals developmental trajectories of Arabidopsis root cells
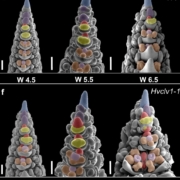
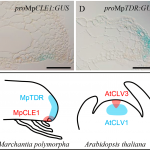
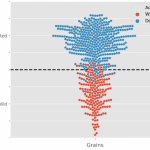
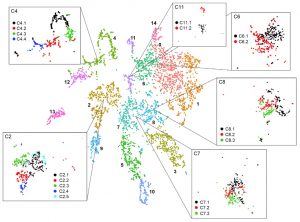 New advances in sequencing technologies have made possible the understanding of the dynamics of developmental processes in an unprecedented manner. In a recent study, Denyer, Ma, et al. profiled nearly 5,000 cells from Arabidopsis roots using single-cell RNA sequencing technology. Gene expression profiles grouped cells into clusters representing the major cell types in the root; each cluster included some well-characterized genes (i.e. PLT1, SCR, SHR), so, it was possible to assign its identity, additionally new cell-type specific genes were identified, for some of them, the predicted expression pattern was confirmed using reporter lines. Furthermore, sub-clustering allowed the identification of cells in a transitional state, so it was possible to follow a developmental trajectory. An example of these was made with the analysis of the transition from stem cells to mature trichoblast. Authors also analyzed clusters derived from cell populations of the shortroot-3 mutant, they detect the expected absence of the endodermis clusters and detected hidden defects in other cell types. This work provides an excellent spatiotemporal perspective of root cell differentiation. (Summary by Humberto Herrera-Ubaldo) Devel. Cell 10.1016/j.devcel.2019.02.022
New advances in sequencing technologies have made possible the understanding of the dynamics of developmental processes in an unprecedented manner. In a recent study, Denyer, Ma, et al. profiled nearly 5,000 cells from Arabidopsis roots using single-cell RNA sequencing technology. Gene expression profiles grouped cells into clusters representing the major cell types in the root; each cluster included some well-characterized genes (i.e. PLT1, SCR, SHR), so, it was possible to assign its identity, additionally new cell-type specific genes were identified, for some of them, the predicted expression pattern was confirmed using reporter lines. Furthermore, sub-clustering allowed the identification of cells in a transitional state, so it was possible to follow a developmental trajectory. An example of these was made with the analysis of the transition from stem cells to mature trichoblast. Authors also analyzed clusters derived from cell populations of the shortroot-3 mutant, they detect the expected absence of the endodermis clusters and detected hidden defects in other cell types. This work provides an excellent spatiotemporal perspective of root cell differentiation. (Summary by Humberto Herrera-Ubaldo) Devel. Cell 10.1016/j.devcel.2019.02.022