Setting and Diffusing the Cyanide Bomb in Plant Defense
Hydrogen cyanide (HCN) is a potent inhibitor of cytochrome c oxidase, a conserved component of the respiratory electron transport chain in all aerobic life. Thus, HCN is well suited to serve as a broad-spectrum chemical defense, and indeed it plays such a role in many interactions between plants and other organisms. However, HCN and its immediate biosynthetic precursors, cyanohydrins, are too reactive to be deployed directly as preformed defenses. Instead, plants have repeatedly evolved a strategy of disarming and sequestering cyanohydrins as glycosylated derivatives, termed cyanogenic glycosides. These compounds can be used as substrates for the rapid production of HCN by the action of glycosidase and α-hydroxynitrilase enzymes. The accumulation of these activating enzymes is compartmentalized from that of their substrates so that they do not mix until the plant has been damaged. Upon damage, however, a chewing insect or other pest quickly discovers that the conflict has escalated to chemical warfare.
Cyanogenic glycosides are found in many plant species, including important crops such as sorghum (Sorghum bicolor), cassava (Manihot esculenta), and Rosaceae species like apple (Malus domestica) and Prunus spp. (stone fruits and almonds [Prunus dulcis]). Structurally, these compounds are defined by their precursor amino acid and glycans. Two widely distributed cyanogenic glycosides are the mono- and diglucosyl derivatives of mandelonitrile: prunasin and amygdalin (Fig. 1). In this issue of Plant Physiology, reports elucidate distinct biosynthetic routes to mandelonitrile and its cyanogenic glucosides in almond (Thodberg et al., 2018) and sugar gum (Eucalyptus cladocalyx; Hansen et al., 2018).
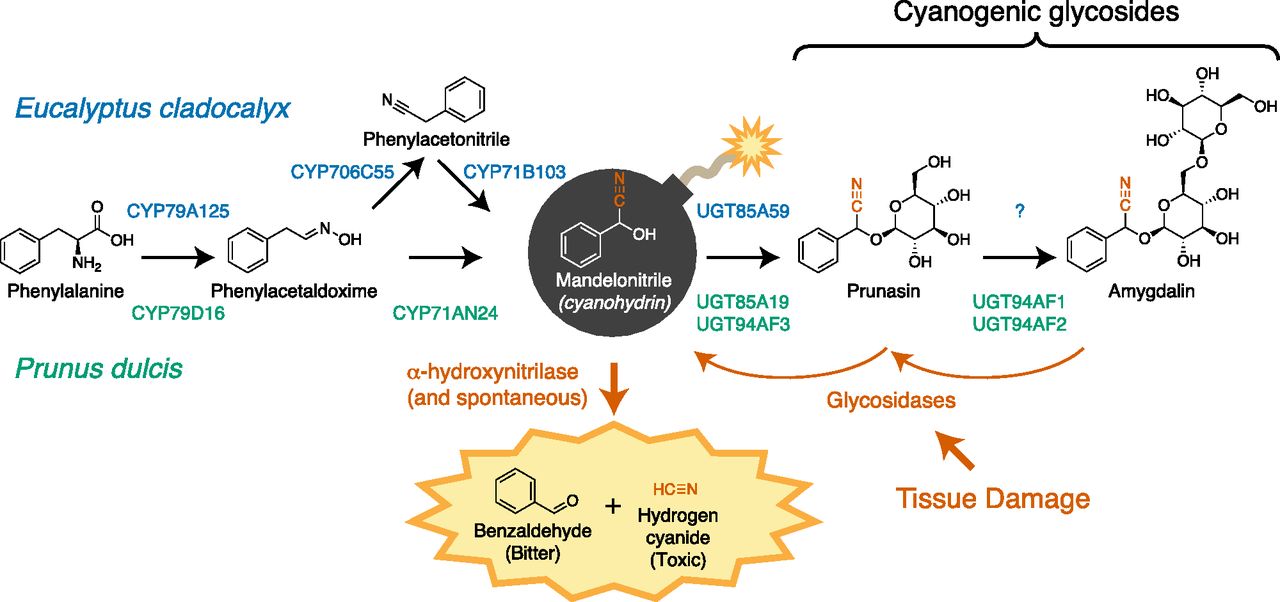
In most Rosaceae crops, prunasin and amygdalin are present in parts of the plant already considered inedible, such as seeds and leaves. In almonds, however, the kernel is of culinary interest, and amygdalin in this tissue can confer unacceptable toxicity and bitterness. In this case, amygdalin accumulation is a Mendelian trait determined by an unknown gene at the Sweet-kernel (Sk) locus. To better understand the genetic basis of bitterness, Thodberg et al. (2018) characterized the amygdalin biosynthesis pathway in almond. While the pathway they identified is largely congruent with previous reports, this is the first report of the enzyme catalyzing the final step of amygdalin synthesis, the glucosylation of prunasin (Fig. 1). This newly reported enzyme, however, does not appear to be the determinant of amygdalin content in bitter almonds. Instead, the authors show that expression of CYP79D16 and CYP71AN24, two genes encoding the cytochrome P450s that yield mandelonitrile from Phe, is strongly repressed in the kernels of sweet almonds that do not accumulate amygdalin. Since the genes encoding these enzymes do not map to the Sk locus, the gene underlying the Sk phenotype likely encodes a transcription factor or other transacting regulator of CYP79D16 and CYP71AN24 expression. This work now sets the stage for identifying these regulators and developing reliable molecular markers for bitterness in almond breeding programs.
Comparative studies have consistently revealed that pathways for specialized metabolite synthesis evolve as variations on themes, with specific components repeatedly recruited from the same clades of large protein families (Pichersky and Lewinsohn, 2011). In the case of cyanogenic glycosides, numerous studies have established a canonical pathway: amino acids are first converted to their respective acetaldoximes by enzymes belonging to the CYP79 family of cytochrome P450 (CYP) enzymes and then converted to a cyanohydrin in a single step by a member of the CYP71 or CYP736 family. Cyanohydrins are then typically glycosylated by UDP-Glc glycosyl transferases (UGTs) from the UGT85 family (Gleadow and Møller, 2014). A second report in this issue of Plant Physiology details the pathway for prunasin biosynthesis in sugar gum and reveals an unexpected reconfiguration of this pathway (Hansen et al., 2018).
To identify the prunasin pathway in sugar gum, Hansen et al. used transcriptome data to find candidate genes from the expected CYP and UGT families. However, initial attempts to reconstitute the pathway by expressing these genes in Nicotiana benthamiana were unsuccessful. Specifically, CYP71B103 was unable to use phenylacetaloxime as a substrate for mandelonitrile production. Although the product of CYP71B103 was mandelonitrile, its substrate was unexpectedly phenylacetonitrile. The existence of this intermediate implies that the bifunctional activity seen in other cyanohydrin-producing CYP71 enzymes is split between two enzymes in sugar gum. The second, nitrile-producing enzyme was found to be CYP706C55, a member of a CYP family that has not been previously implicated in cyanogenic glycoside synthesis. Thus, sugar gum demonstrates a variation on the theme of CYP and UGT recruitment to specialized metabolite biosynthesis. Notably, this more complex pathway is still able to accommodate a substantial biosynthetic flux to cyanogenic glycosides, which can account for 20% of nitrogen utilization in leaves of this species.
New genomics and analytical technologies allow unprecedented opportunities to explore the evolution and diversity of natural product biosynthesis, but interpretation of these new data rely on foundational knowledge painstakingly acquired via classical biochemistry. The work of Thodberg et al. and Hansen et al. provides an opportunity to celebrate the life and work of Professor Eric Conn, a pioneer of plant biochemistry, who passed away last year. Eric Conn and his collaborators are responsible for elucidating many of the principles of plant cyanogenesis, including the important finding that cyanogenic glycosides and their hydrolytic enzymes are sequestered at the cellular and subcellular level until tissue damage occurs (Conn, 2008). These insights established cyanogenic glycosides as models for other conjugated chemical defense compounds, such as glucosinolates, which are similarly segregated from their activating myrosinase enzymes (Lüthy and Matile, 1984). The study of plant metabolism owes much to Eric Conn’s groundbreaking work, and the continued progress in this field reflects Isaac Newton’s sentiment that “If I have seen further it is by standing on the shoulders of Giants.”
REFERENCES
Conn EE (2008) Our work with cyanogenic plants. Annu Rev Plant Biol 59: 1–19
Gleadow RM, Møller BL (2014) Cyanogenic glycosides: synthesis, physiology, and phenotypic plasticity. Annu Rev Plant Biol 65: 155–185
Lüthy B, Matile P (1984) The mustard oil bomb: rectified analysis of the subcellular organisation of the myrosinase system. Biochem Physiol Pflanz 179: 5–12
Pichersky E, Lewinsohn E (2011) Convergent evolution in plant specialized metabolism. Annu Rev Plant Biol 62: 549–566