SAUR15 connects auxin perception to lateral and adventitious root formation
Charles Copeland1
1Department of Plant Microbe Interactions, Max Planck Institute for Plant Breeding Research, 50829 Cologne, Germany
ORCID: 0000-0001-8535-8615
As plants grow, they produce new organs such as leaves, lateral roots, and flowers. Plant development is regulated by complex interactions of plant hormones and other signaling molecules, allowing plants to modulate their growth according to multiple environmental cues. Auxin is one of the major hormones controlling plant development. AUXIN RESPONSE FACTOR (ARF) transcription factors are de-repressed upon auxin perception, and initiate downstream signaling pathways. Within minutes of auxin treatment, the transcription of a large family of plant-specific genes called the SAURs is rapidly induced (Ren and Gray, 2015). The rapid nature of this induction suggests that SAUR genes may be direct targets of ARFs, and indeed, the promoters of many SAUR genes contain predicted ARF binding sites. SAURs are necessary for many auxin-mediated responses, particularly growth via cell elongation. However, due to the large number of SAURs and their complex layers of redundancy and specificity, the roles of individual members have not been well characterized. For example, although Arabidopsis thaliana SAUR15 has been used as a marker to study the details of auxin-induced gene expression (Walcher and Nemhauser, 2012), the physiological role of SAUR15 was unknown.
 In this issue of Plant Physiology, co-first authors Hongju Yin and Mengzhan Li et al. (2020) report a genetic and biochemical function of SAUR15 in auxin signaling. They show that SAUR15 works downstream of ARFs to promote cell expansion, and that this function is especially important for lateral root development. First, Yin, Li et al. (2020) examined the phenotype of SAUR15 overexpression (SAUR15-OE) and saur15 mutant plants. These lines show contrasting phenotypes for a number of auxin-related growth characteristics, such as root architecture. SAUR15-OE plants have longer roots with more lateral root branches, while saur15 mutants have shorter roots and fewer lateral roots. Etiolated SAUR15-OE plants also develop more adventitious roots, which are roots that form from non-root tissue. The developmental steps in lateral root formation have been relatively well characterized, and auxin signaling appears to be critical at each step (Atkinson et al., 2014). For example, auxin-mediated cell expansion is required for the formation of lateral root primordia and for emergence of the lateral roots through the outer layers of the primary root. Adventitious root formation appear to follow a similar pattern as lateral root formation, and is also reliant on auxin (Atkinson et al., 2014). Thus, the authors hypothesized that SAUR15 acts downstream of auxin to promote lateral and adventitious root development.
In this issue of Plant Physiology, co-first authors Hongju Yin and Mengzhan Li et al. (2020) report a genetic and biochemical function of SAUR15 in auxin signaling. They show that SAUR15 works downstream of ARFs to promote cell expansion, and that this function is especially important for lateral root development. First, Yin, Li et al. (2020) examined the phenotype of SAUR15 overexpression (SAUR15-OE) and saur15 mutant plants. These lines show contrasting phenotypes for a number of auxin-related growth characteristics, such as root architecture. SAUR15-OE plants have longer roots with more lateral root branches, while saur15 mutants have shorter roots and fewer lateral roots. Etiolated SAUR15-OE plants also develop more adventitious roots, which are roots that form from non-root tissue. The developmental steps in lateral root formation have been relatively well characterized, and auxin signaling appears to be critical at each step (Atkinson et al., 2014). For example, auxin-mediated cell expansion is required for the formation of lateral root primordia and for emergence of the lateral roots through the outer layers of the primary root. Adventitious root formation appear to follow a similar pattern as lateral root formation, and is also reliant on auxin (Atkinson et al., 2014). Thus, the authors hypothesized that SAUR15 acts downstream of auxin to promote lateral and adventitious root development.
Lateral and adventitious roots are critically important for plant growth. A branched root system provides structural support by anchoring the plant to the substrate, and increasing the surface area available for water and nutrient scavenging. Plants regulate their root architecture in response to environmental cues. For example, lateral root formation is upregulated in soil patches with higher moisture content, and suppressed in drier patches, in order to maximize water uptake (Maurel and Nacry, 2020). Phosphate starvation also triggers increased lateral root formation in shallow soil layers, as plant-accessible phosphate is often more abundant near the soil surface (Atkinson et al., 2014). Recently, aerial adventitious roots from indigenous maize varieties were reported to host nitrogen-fixing bacteria, reducing the plant’s reliance on nitrogen from the soil (Van Deynze et al., 2018). Given the importance of lateral and adventitious roots, increased knowledge about their formation should improve our understanding of how plants respond to varying soil environments, and may provide opportunities for crop improvement.
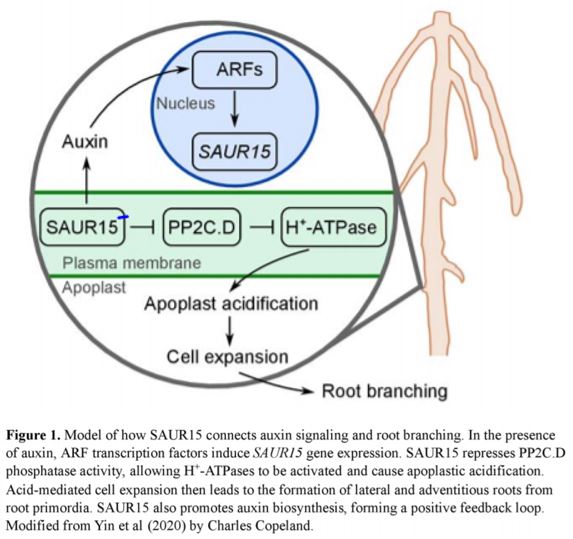
Using genetic and biochemical methods, the authors examined how SAUR15 connects auxin perception and root branching. They found that arf6 and arf7 mutants have reduced SAUR15 expression and fewer lateral roots, but overexpression of SAUR15 in these mutants restores the number of lateral and adventitious roots. Chromatin immunoprecipitation followed byquantitative PCR (ChIP-qPCR) showed that ARF6 and ARF7 bind directly to the promoter of SAUR15. This indicates that ARF transcription factors promote root branching by inducing SAUR15 expression.
The authors next studied the downstream action of SAUR15, using knowledge from different SAUR proteins as a guide. A number of other SAURs interact with and inhibit PP2C.D phosphatases, which in turn allows for activation of plasma membrane-localized H+-ATPases such as AHA1 (Ren and Gray, 2015; Wong et al., 2019). H+-ATPase activity results in acidification of the apoplast and subsequent loosening of the cell wall, allowing cells to expand (Ren and Gray, 2015; Barbez et al., 2017).
Although apoplast acidification by H+-ATPases has been shown to be a major mechanism of auxin-mediated cell expansion during root and shoot growth, its role in root branching had not yet been reported. The results obtained by Yin et al. (2020) support the idea that SAUR15 also uses this pathway to promote lateral and adventitious root formation, as described in Fig. 1. The reduced lateral root production of saur15 mutants is suppressed in saur15 pp2c.d double mutants and in saur15 AHA1-OE lines. In addition, saur15 mutants show decreased H+-ATPase activity and correspondingly smaller drop in apoplastic pH, which likely reduces the ability of the cells to expand.
Besides repressing PP2C.D, SAUR15 also appears to play an second role in lateral and adventitious root formation by promoting auxin biosynthesis (Yin et al., 2020). Increased auxin abundance could initiate a positive feedback loop, promoting lateral root production through accumulation of SAUR proteins and other auxin-responsive proteins. It is not known how SAUR15, which localizes to the plasma membrane, can induce auxin biosynthesis. SAUR proteins share a well-conserved central domain, but their protein structures do not provide any obvious clues about their biochemical function (Ren and Gray, 2015; Stortenbeker and Bemer, 2019). However, the N- and C-termini of SAUR proteins are more variable and have been proposed to interact with different proteins or signaling molecules.
Yin et al (2020) identified SAUR15 as an important intermediary between auxin perception and root branching. Interestingly, SAUR15 expression is also regulated by brassinosteroids and possibly other plant hormones (Walcher and Nemhauser, 2012; Yin et al., 2020). This suggests that SAUR15 could serve a point of crosstalk between different hormone signaling pathways. Future research could focus on whether SAUR15 regulates lateral and adventitious root growth in response to environmental conditions.
Literature Cited
Atkinson JA, Rasmussen A, Traini R, Voß U, Sturrock C, Mooney SJ, Wells DM, Bennett MJ (2014) Branching out in roots: Uncovering form, function, and regulation. Plant Physiol 166: 538–550
Barbez E, Dünser K, Gaidora A, Lendl T, Busch W (2017) Auxin steers root cell expansion via apoplastic pH regulation in Arabidopsis thaliana . Proc Natl Acad Sci 114: E4884–E4893
Van Deynze A, Zamora P, Delaux PM, Heitmann C, Jayaraman D, Rajasekar S, Graham D, Maeda J, Gibson D, Schwartz KD, et al (2018) Nitrogen fixation in a landrace of maize is supported by a mucilage-associated diazotrophic microbiota. PLoS Biol 16: 1–21
Maurel C, Nacry P (2020) Root architecture and hydraulics converge for acclimation to changing water availability. Nat Plants. doi: 10.1038/s41477-020-0684-5
Ren H, Gray WM (2015) SAUR Proteins as Effectors of Hormonal and Environmental Signals in Plant Growth. Mol Plant 8: 1153–1164
Stortenbeker N, Bemer M (2019) The SAUR gene family: The plant’s toolbox for adaptation of growth and development. J Exp Bot 70: 17–27
Walcher CL, Nemhauser JL (2012) Bipartite promoter element required for auxin response. Plant Physiol 158: 273–282
Wong JH, Spartz AK, Park MY, Du M, Gray WM (2019) Mutation of a conserved motif of PP2C.D phosphatases confers SAUR immunity and constitutive activity. Plant Physiol 181: 353–366