RNA Degradome Studies Give Insights into Ribosome Dynamics
IN BRIEF by Gregory Bertoni [email protected]
RNA metabolism is key to a number of crucial processes in the cell, including transcription, RNA splicing, translation, and gene regulation. For efficient translation, mature mRNAs must have a 7-methylguanosine cap on the 5′ end to help recruit the translation machinery and protect the mRNA from exonucleases. If the cap is removed during RNA degradation, a free 5′ monophosphate is revealed. Likewise, when an endonuclease cleaves an RNA molecule that is a microRNA (miRNA) or small interfering RNA target, it also leaves a free 5′ monophosphate. Therefore, RNA degradation products (the RNA degradome) can be identified by sequencing RNAs that have a free 5′ monophosphate.
Transcriptome-wide methods to survey the RNA degradome were first developed in plants to inventory miRNA and small interfering RNA targets (Addo-Quaye et al., 2008; German et al., 2008; Gregory et al., 2008). Because the translating ribosome protects the underlying mRNA from degradation, analysis of the RNA degradome can also provide insights into ribosome positioning and movement during translation (Pelechano et al., 2015). This analysis can also identify sites of protein binding and miRNA-induced cleavage in RNA molecules and can verify the translatability of upstream open reading frames (uORFs) that are often found in the 5′ untranslated regions of genes (Rogers and Chen, 2013; von Arnim et al., 2014).
Two articles in this issue of The Plant Cell analyze the cotranslational RNA degradome by isolating poly(A)+ RNAs having a free 5′ phosphate and identifying and quantifying the nucleotide positions of the free 5′ end. Both research groups found a 3-nucleotide periodicity in the size of RNA degradation products corresponding to coding regions, providing physical evidence that the ribosome moves in 3-nucleotide increments during translation. In addition, they found a 30-nucleotide periodicity in the size distribution of RNA degradome fragments, a distance that corresponds to the amount of mRNA covered by a single translating ribosome.
Yu et al. (2016) used genome-wide mapping of uncapped and cleaved transcripts (GMUCT; Willmann et al., 2014) to study the RNA degradome in Arabidopsis thaliana leaves and unopened flower buds. In the GMUCT procedure, poly(A)+ RNA is isolated and an adaptor is immediately ligated to all free 5′ monophosphate ends, which excludes mature capped mRNAs and therefore represents mRNA degradation products. These RNAs are then purified and used in reverse transcription reactions to produce a library of RNAs representing all uncapped and cleaved mRNAs in a transcriptome. After sequencing, the positions and abundances of the 5′ ends of these RNA fragments can be determined and quantified.
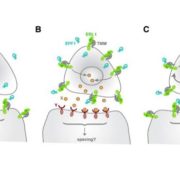
In addition to the 3-nucleotide periodicity in cleavage sites in the coding regions of mRNAs, Yu et al. discovered an accumulation of cleavage sites 16 to 17 nucleotides upstream of stop codons, presumably representing ribosomal pausing during translation termination (see figure, top panel). Interestingly, they observed a slightly different profile of 5′ read ends depending on whether the stop codon was TAA, TGA, or TAG. The authors suggest that this represents differential accessibility to 5′ to 3′ exoribonucleases at the 5′ boundary of pausing ribosomes.
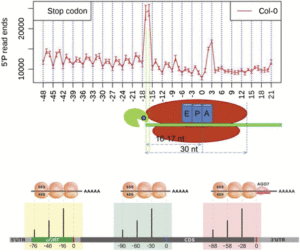
Treatment of plants with cycloheximide, an inhibitor of translation elongation, resulted in a preponderance of cleavage events 13 to 14 nucleotides upstream of the start codon, representing RNA cleavage adjacent to ribosomes stalled at the start codon. The researchers also found peaks of cleavage events in the 5′ untranslated regions of some transcripts, located 16 to 17 nucleotides upstream of the end of uORFs found in these untranslated regions. They also found a peak at 46 to 47 nucleotides upstream of the end of the uORF, and because a ribosome covers ∼30 nucleotides during translation, this suggests the presence of tandem ribosomes paused and stacked up at the stop codon, giving further evidence that these uORFs are actually translated.
Next, Yu et al. used xrn4 mutant plants to show the involvement of the exoribonuclease XRN4 in cotranslational decay of mRNA. In xrn4 mutant plants, there was no evidence of the 3-nucleotide periodicity of cleavage sites and there also was no enrichment of cleavage sites at the ribosome boundary near the stop codon. They then showed that the mRNA cap-binding complex, which helps regulate mRNA stability, spicing, and processing, was also involved in cotranslational RNA decay.
Using a similar strategy known as parallel analysis of RNA ends (German et al., 2008), Hou et al. (2016) found a 3-nucleotide periodicity in the distribution of free 5′ mRNA ends not only in Arabidopsis but also in rice (Oryza sativa) and soybean (Glycine max), with a bias toward the frame used in translation. They not only found an accumulation of cleavage events 16 nucleotides upstream of the stop codon, but also found an additional 30-nucleotide periodicity in cleavage sites, noting peaks at −46 and −76, suggesting that ribosomes can be stacked up adjacently in pairs and in threes during translation termination.
By reverse searching for uORFs with an accumulation of 5′ RNA ends peaking 16 or 46 nucleotides upstream of the stop codon, Hou et al. were able to identify potential regulatory uORFs. In this search, they found four known and four novel uORFs with the presumed ability to stall ribosomes. One uORF was found in CBL-INTERACTING PROTEIN KINASE6, a member of a protein family that interacts with calcineurin B-like proteins to mediate plant responses to stresses such as pathogen challenge or high salt. They then provide evidence that ribosome stalling in this uORF provides a means to regulate the expression of downstream coding sequences. They also discovered potential ribosome stalling at uORFs in MYB34 and MYB51, which are MYB transcription factors involved in the regulation of glucosinolate biosynthesis
Hou et al. discovered an accumulation of 5′ RNA ends in the vicinity of motifs recognized by RNA binding proteins, suggesting that their attachment to target RNA sequences would protect them from degradation. They analyzed the pattern of 5′ RNA ends and ribosome footprints to study whether plant miRNAs can directly block the movement of ribosomes. By analyzing the RNA degradome of three Arabidopsis TAS genes, which produce trans-acting silencing RNAs, they not only provided evidence that a miR390-AGO7 complex arrests a ribosome, but also were able to discriminate whether the ribosome stalling was during translation elongation or termination.
Additional studies investigated a small group of bZIP genes, which regulate the translation of downstream ORFs in response to sucrose concentration. We look forward to future studies that analyze the RNA degradome and anticipate further insights into the mechanisms of cotranslational RNA decay, uORF regulatory mechanisms, and ribosome dynamics during translation.
REFERENCES
Addo-Quaye, C., Eshoo, T.W., Bartel, D.P., and Axtell, M.J. (2008). Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 18: 758–762.
German, M.A., et al. (2008). Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 26: 941–946.
Gregory, B.D., O’Malley, R.C., Lister, R., Urich, M.A., Tonti-Filippini, J., Chen, H., Millar, A.H., and Ecker, J.R. (2008). A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell 14: 854–866.
Hou, C.-Y., Lee, W.-C., Chou, H.-C., Chen, A.-P., Chou, S.-J., and Chen, H.-M. (2016). Global analysis of truncated RNA ends reveals new insights into ribosome stalling in plants. Plant Cell 28: 2398–2416.
Pelechano, V., Wei, W., and Steinmetz, L.M. (2015). Widespread co-translational RNA decay reveals ribosome dynamics. Cell 161: 1400–1412.
Rogers, K., and Chen, X. (2013). Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25: 2383–2399.
Willmann, M.R., Berkowitz, N.D., and Gregory, B.D. (2014). Improved genome-wide mapping of uncapped and cleaved transcripts in eukaryotes–GMUCT 2.0. Methods 67: 64–73.
Yu, X., Willmann, M.R., Anderson, S.J., and Gregory, B.D. (2016). Genome-wide mapping of uncapped and cleaved transcripts reveals a role for the nuclear mRNA cap-binding complex in contranslational RNA decay in Arabidopsis. Plant Cell 28: 2385–2397.
von Arnim, A.G., Jia, Q., and Vaughn, J.N. (2014). Regulation of plant translation by upstream open reading frames. Plant Sci. 214: 1–12.





Leave a Reply
Want to join the discussion?Feel free to contribute!