Review: DNA sequencing at 40: past, present and future ($)
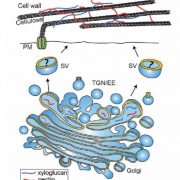
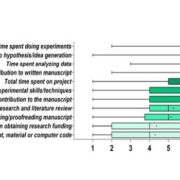
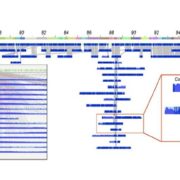
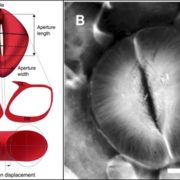
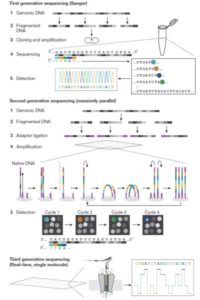 Shendure et al. provide a superb review of how DNA sequencing technology has changed over the years and how these changes open up new applications. They start with the Maxam and Gilbert chemical cleavage and the Sanger “chain-termination” methods developed in the 1970s, and describe the scale-ups required for the draft completion in 2001 of the Human Genome Project. These “first-generation” approaches were followed by the development of massively parallel or “next-generation” and “sequence-by-synthesis” methods in the 2000s, now dominated by the Illumina platform. We’re now in the era of “real-time, single-molecule” sequencing, exemplified by PacBio and Nanopore technologies. Although these single-molecule long reads are still prone to errors, they provide the essential longer scaffolds to which short reads can be aligned. This excellent review includes a timeline of genome milestones [including our favs, Arabidopsis thaliana (2000), Oryza sativa (2005), Cyanidioschyzon merolae (a red alga, 2007) and Zea mays (2009)] as well as computational and application milestones. The review ends wioth a look at the future technologies and applications of DNA sequencing, which the authors predict “will have a longevity and impact on par with or exceeding that of the microscope.” A super review article for teaching! Nature 10.1038/nature24286
Shendure et al. provide a superb review of how DNA sequencing technology has changed over the years and how these changes open up new applications. They start with the Maxam and Gilbert chemical cleavage and the Sanger “chain-termination” methods developed in the 1970s, and describe the scale-ups required for the draft completion in 2001 of the Human Genome Project. These “first-generation” approaches were followed by the development of massively parallel or “next-generation” and “sequence-by-synthesis” methods in the 2000s, now dominated by the Illumina platform. We’re now in the era of “real-time, single-molecule” sequencing, exemplified by PacBio and Nanopore technologies. Although these single-molecule long reads are still prone to errors, they provide the essential longer scaffolds to which short reads can be aligned. This excellent review includes a timeline of genome milestones [including our favs, Arabidopsis thaliana (2000), Oryza sativa (2005), Cyanidioschyzon merolae (a red alga, 2007) and Zea mays (2009)] as well as computational and application milestones. The review ends wioth a look at the future technologies and applications of DNA sequencing, which the authors predict “will have a longevity and impact on par with or exceeding that of the microscope.” A super review article for teaching! Nature 10.1038/nature24286