Proliferate at Your Own Risk: Ribosomal Stress and Regeneration
Plant growth and development are extremely adaptable to changes in the external environment, including nutrient status, light quality and intensity, and temperature. Thanks to their developmental plasticity, plants can also initiate new organs from differentiated tissue following wounding, to the benefit of horticulturists everywhere.
Plants establish a balance between cell proliferation and cell growth that relies heavily on protein synthesis by ribosomes, so it is not surprising that mutants affected in ribosome biogenesis or function show pleiotropic growth defects, including loss of tissue regeneration. New results from M. Sugiyama and colleagues identify the NAC domain transcription factor ANAC082 as the effector of a surveillance mechanism monitoring ribosomal health (Ohbayashi et al., 2017).
Sugiyama and colleagues invested considerable time and effort to generate a collection of temperature-sensitive mutants that would allow a genetic dissection of tissue regeneration in Arabidopsis (reviewed by Sugiyama, 2014). The advantage of such mutants is obvious here, as they conditionally bypass pleiotropy or seedling lethality. Root or hypocotyl explants from individual mutant lines are challenged for regeneration potential of calli, roots, or shoots by modulating the balance of the growth hormones auxin and/or cytokinin.
The first hint that regeneration requires ribosome activity came about with the isolation of the shoot redifferentiation defective 2 (srd2) mutant (Ohtani and Sugiyami, 2005). SRD2 encodes a protein similar to human SNAP50 (snRNA-activating protein). The SNAP complex recognizes a motif in the promoter of small nuclear RNAs (snRNAs) U1, U2, and U6, all involved in mRNA splicing, and recruits RNA polymerase II or III (depending on the gene) to initiate transcription. The expression of several snRNAs is induced during callus regeneration at 22ºC and 28ºC, but not in the mutant. Levels of the small nucleolar RNA U3 (U3 snoRNA), thought to participate in ribosomal RNA pre-processing, are also reduced. The U3 snoRNA results set the stage for the shape of things to come.
The cloning of ROOT INITIATION DEFECTIVE 2 (RID2) consolidated the connection between regeneration and ribosomes (Ohbayashi et al., 2011). RID2 loss-of-function mutants are blocked in the initiation of root primordia and have unusually swollen nucleoli. This suggests a defect in ribosome biogenesis, and in fact, rid2 mutants accumulate excessive amounts of intermediates during pre-ribosomal RNA processing.
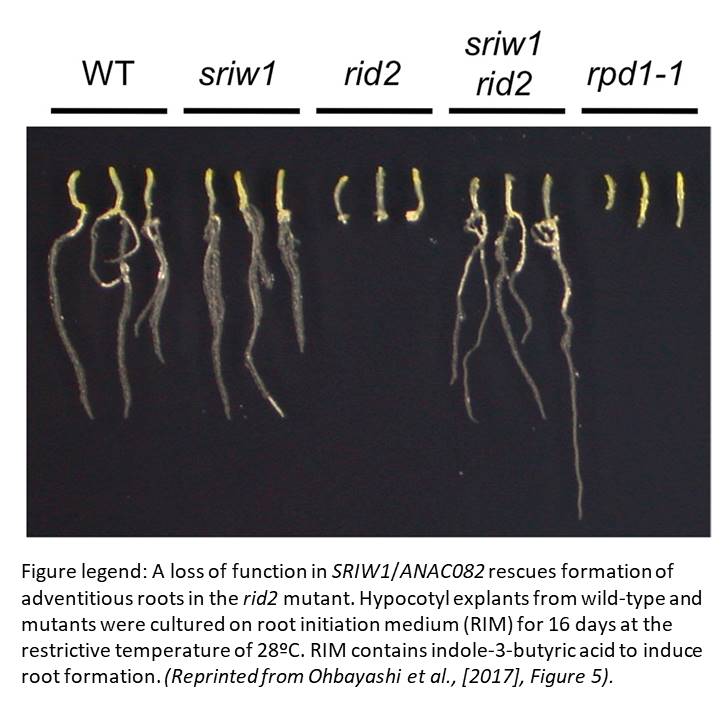
RID2 null mutant alleles are female gametophyte-lethal or embryo-lethal, thus validating the temperature-sensitive screen approach. In addition, the initial rid2-1 mutant is a weak allele, which makes for an ideal background for a suppressor screen and brings us to the isolation of sriw1/anac082. SRIW1 encodes the NAC-domain transcription activator ANAC082. sriw1 mostly suppresses phenotypes seen in rid2-1 and several other ribosome biogenesis-related mutants, including root length, leaf blade size, venation pattern and regeneration potential, but not the accumulation of rRNA processing intermediates.
In animals, ribosome function can be disturbed by stress, including starvation, hypoxia, pharmacological agents, or genetic defects. Activation of the ribosome stress response results in cell-cycle arrest or programmed cell death and involves the tumor suppressor gene p53. In a possible parallel with p53, ribosome-related stress in Arabidopsis induces ANAC082 expression and blocks tissue regeneration (Ohbayashi et al., 2017).
What are the signals that stimulate ANAC082 expression following ribosome-related stress, and what are the targets of ANAC082 that mediate cell cycle arrest? Interestingly, the promoter of ANAC082 possesses an upstream open reading frame (uORF). Ribosome stress might decrease translation of this uORF and alleviate some of the imposed transcriptional repression of the downstream gene, which may provide a mechanism to adjust ANAC082 expression where the watcher is, in fact, being watched.
REFERENCES
Ohbayashi I, Konishi M, Ebine K, Sugiyama M (2011). Genetic identification of Arabidopsis RID2 as an essential factor involved in pre-rRNA processing. Plant J. 2011 67: 49-60.
Ohbayashi I, Lin C-Y, Shinohara N, Matsumura Y, Machida Y, Horiguchi G, Tsukaya H, Sugiyama M (2017). Evidence for a Role of ANAC082 as a Ribosomal Stress Response Mediator Leading to Growth Defects and Developmental Alterations in Arabidopsis. Plant Cell, published September 12 2017. https://doi.org/10.1105/tpc.17.00255
Ohtani M and Sugiyama M (2005). Involvement of SRD2-mediated activation of snRNA transcription in the control of cell proliferation competence in Arabidopsis. Plant J. 43: 479-90.
Sugiyama, M. (2014) Molecular genetic analysis of organogenesis in vitro with temperature-sensitive mutants. Plant Biotech. Rep. 8: 29-34.