POME: Quantitative and dynamic cell polarity tracking pipeline (bioRxiv)
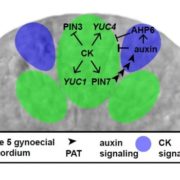
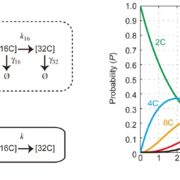
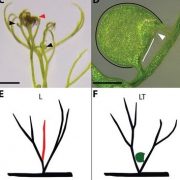
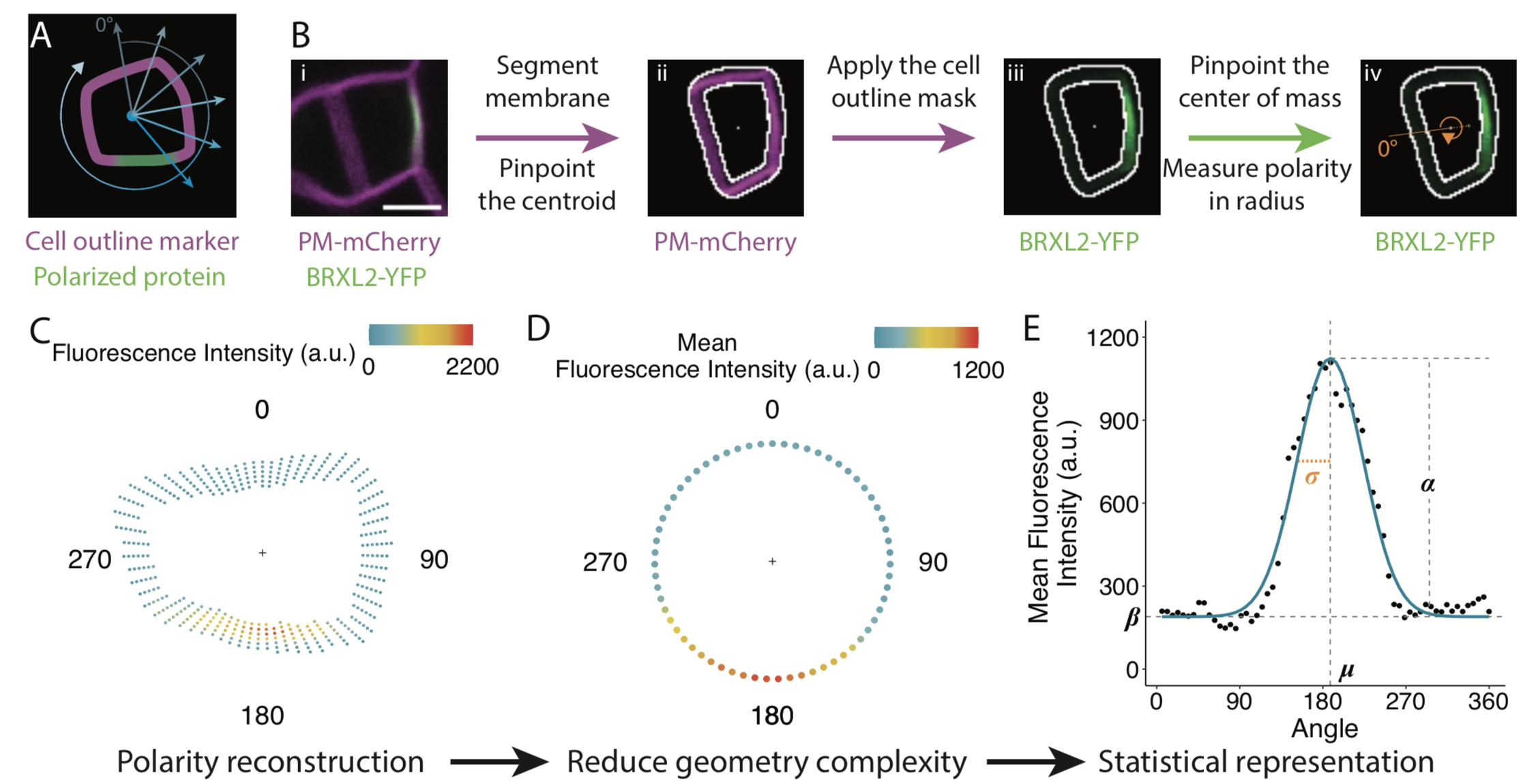 Many proteins polarize in the cell creating a subcellular niche for various functions. Asymmetric distribution of proteins is a general mechanism for localized growth, directional long-range signaling, cell migration, and asymmetric cell divisions. Well-known examples of polarity proteins include PIN-FORMED1 in auxin transport, and BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE (BASL), and BREVIS RADIX family (BRXf) in stomatal asymmetric cell divisions. Most data on polarized proteins are qualitative, without much information on protein quantity, size, and dynamics of the polar domain. In this paper, Gong et al. developed the POME (Polarity Mediated) pipeline for quantitative data analysis of polar proteins with good statistical power. POME is a semiautomated tool that requires a confocal image taken with two different channels (one must highlight the outline of the cell), the FIJI macro, and R script. The macro extracts the cell outline and determines both the amplitude and size of the polarity distribution. The authors have validated the POME using known polarized and depolarized proteins and show that BRXL2 accumulates prior to BASL in Arabidopsis. The authors also extended this analysis to other cell types (root cells) and model organisms (Brachypodium, C. elegans) to determine protein polarity. Thus, POME can be used in diverse systems to get quantitative information of polarity with statistical confidence. (Summary by Vijaya Batthula @Vijaya_Batthula) bioRxiv. 10.1101/2020.09.12.294942.
Many proteins polarize in the cell creating a subcellular niche for various functions. Asymmetric distribution of proteins is a general mechanism for localized growth, directional long-range signaling, cell migration, and asymmetric cell divisions. Well-known examples of polarity proteins include PIN-FORMED1 in auxin transport, and BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE (BASL), and BREVIS RADIX family (BRXf) in stomatal asymmetric cell divisions. Most data on polarized proteins are qualitative, without much information on protein quantity, size, and dynamics of the polar domain. In this paper, Gong et al. developed the POME (Polarity Mediated) pipeline for quantitative data analysis of polar proteins with good statistical power. POME is a semiautomated tool that requires a confocal image taken with two different channels (one must highlight the outline of the cell), the FIJI macro, and R script. The macro extracts the cell outline and determines both the amplitude and size of the polarity distribution. The authors have validated the POME using known polarized and depolarized proteins and show that BRXL2 accumulates prior to BASL in Arabidopsis. The authors also extended this analysis to other cell types (root cells) and model organisms (Brachypodium, C. elegans) to determine protein polarity. Thus, POME can be used in diverse systems to get quantitative information of polarity with statistical confidence. (Summary by Vijaya Batthula @Vijaya_Batthula) bioRxiv. 10.1101/2020.09.12.294942.