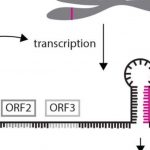
Peptide-Receptor Signaling Pumps the Brakes on Auxin Biosynthesis and Ethylene Signaling to Harmonize Root Growth and Nodulation
Nitrogen (N) is the most abundant element in Earth’s atmosphere. However, plants must capture this essential element from soil through their roots. To do this, legume roots forge symbioses with rhizobia to initiate nodule development. Root nodules provide rhizobia an environment suitable for converting N2 into ammonia. When N is limiting, legumes must balance energetic costs associated with producing lateral roots and nodules. Some regulators of root growth and nodulation are known, such as signaling peptides, leucine-rich repeat–receptor-like kinases (LRR-RLKs), and the phytohormones auxin and ethylene. The mechanisms that harmonize their development remain largely unknown. Here, Zhu and coworkers (2020) identify a molecular brake on auxin biosynthesis and ethylene signaling that is key to coordinating root growth and symbiotic nodulation.
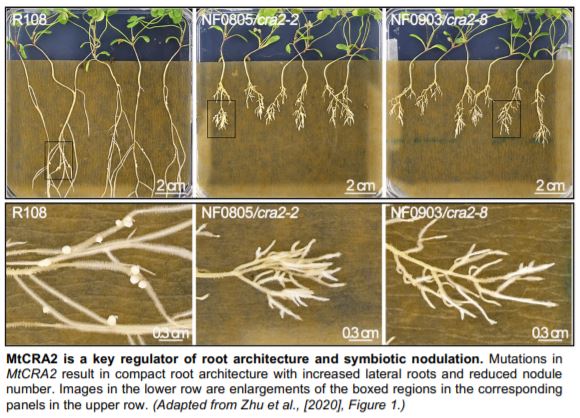 The authors harnessed forward genetics in Medicago truncatula to screen for plants with increased lateral roots and fewer nodules. They identified mutant alleles of the previously reported Compact Root Architecure2 (MtCRA2) encoding LRR-RLK (Huault et al., 2014). Using RNA sequencing, the authors probed downstream targets of the MtCRA2 receptor. Among the nearly 3,000 differentially expressed genes, MtYUCCA2 (MtYUC2) was upregulated in cra2 roots. As MtYUC2 encodes an auxin biosynthesis enzyme, the authors hypothesized its misexpression in cra2 roots may result in an imbalance between lateral root and nodule densities. Indeed, treating plants with the synthetic auxin 1-Naphthaleneacetic acid phenocopied the dense root architecture typified by cra2, as did MtYUC2 overexpression. Conversely, blocking MtYUC activity with 4-biphenyl boronic acid in the cra2 background and CRISPR/Cas9 knockout of MtYUC2 suppressed cra2 root phenotypes. The authors showed MtYUC2 expression could be suppressed by either low N or exogenous application of MtCEP1 (C-terminally Encoded Peptide1), and MtCEP1 expression could be induced by N starvation. Thus, when N is limited, pumping the brake on auxin levels through a MtCEP1-MtCRA2-MtYUC2 module maintains normal root architecture. What factor(s), then, are responsible for balancing nodulation?
The authors harnessed forward genetics in Medicago truncatula to screen for plants with increased lateral roots and fewer nodules. They identified mutant alleles of the previously reported Compact Root Architecure2 (MtCRA2) encoding LRR-RLK (Huault et al., 2014). Using RNA sequencing, the authors probed downstream targets of the MtCRA2 receptor. Among the nearly 3,000 differentially expressed genes, MtYUCCA2 (MtYUC2) was upregulated in cra2 roots. As MtYUC2 encodes an auxin biosynthesis enzyme, the authors hypothesized its misexpression in cra2 roots may result in an imbalance between lateral root and nodule densities. Indeed, treating plants with the synthetic auxin 1-Naphthaleneacetic acid phenocopied the dense root architecture typified by cra2, as did MtYUC2 overexpression. Conversely, blocking MtYUC activity with 4-biphenyl boronic acid in the cra2 background and CRISPR/Cas9 knockout of MtYUC2 suppressed cra2 root phenotypes. The authors showed MtYUC2 expression could be suppressed by either low N or exogenous application of MtCEP1 (C-terminally Encoded Peptide1), and MtCEP1 expression could be induced by N starvation. Thus, when N is limited, pumping the brake on auxin levels through a MtCEP1-MtCRA2-MtYUC2 module maintains normal root architecture. What factor(s), then, are responsible for balancing nodulation?
Ethylene signals through the positive regulator ETHYLENE INSENSITIVE2 (EIN2) to inhibit rhizobia infection (Penmesta and Cook, 1997), and it has been hypothesized MtCEP1-MtCRA2 dampens ethylene signaling to promote nodulation. The big question then became what is the relationship between MtCEP1-MtCRA2 and MtEIN2? The authors first showed nodulation could be partially recovered in cra2 by knocking out MtEIN2 with CRISPR/Cas9. Then authors examined if MtCEP1 could induce MtCRA2 kinase activity—it did, both in vitro and in vivo. Using quantitative proteomics, the authors probed for target proteins in roots phosphorylated by the MtCRA2 LRR-RLK. Surprisingly, amongst the 1,353 unique phosphopeptides from 872 phosphoproteins, two were MtEIN2 peptides. Zhu and coworkers (2020) went on to demonstrate MtCRA2 and MtEIN2 co-localize in the endoplasmic reticulum, and MtCRA2 could trans-phosphorylate MtEIN2. Phosphorylation of EIN2’s C-terminal end blocks its cleavage and prevents it from signaling to the nucleus (Qiao et al., 2012). Importantly, the authors established that cleavage of MtEIN2’s C-terminus could be repressed when N was limited or by exogenous application of MtCEP1. Therefore, low N serves as another brake, this time on ethylene signaling through MtCEP1-MtCRA2-MtEIN2 to promote nodulation.
Zhu and coworkers (2020) summarize their insightful study in a working model, yet some questions remain to be answered. How exactly does MtCRA2 regulate MtYUC2 expression? Are there MtYUC-independent pathways that regulate root growth downstream of MtCRA2? What are the mechanisms by which secreted signaling peptides activate ER-localized LRR-RLKs? Future, detailed investigations on the topic will undoubtedly deepen our understanding by addressing the open and compelling questions raised by this important work.
Josh Strable
Plant Biology Section, School of Integrative Plant Science
Cornell University
ORCID: 0000-0002-0260-8285
REFERENCES
Huault, E., Laffont, C., Wen, J., Mysore, K.S., Ratet, P., Duc, G., and Frugier, F. (2014). Local and Systemic Regulation of Plant Root System Architecture and Symbiotic Nodulation by a Receptor-like Kinase. PLoS Genet. 10: e1004891.
Penmetsa, R.V., and Cook, D.R. (1997). A Legume Ethylene-Insensitive Mutant Hyperinfected by Its Rhizobial Symbiont. Science 275: 527-530.
Qiao, H., Shen, Z., Huang, S.C., Schmitz, R.J., Urich, M.A., Briggs, S.P., and Ecker, J.R. (2012). Processing and Subcellular Trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338: 390-393.
Zhu., F., Deng, J., Chen, H., Liu, P., Zheng, L., Ye, Q., Li, R., Brault, M., Wen, J., Frugier, F., Dong, J., and Wang, T. (2020). A CEP Peptide Receptor-like Kinase Regulates Auxin Biosynthesis and Ethylene Signaling to Coordinate Root Growth and Symbiotic Nodulation. DOI: https://doi.org/10.1105/tpc.19.00428