Division of labor during apical hook formation
Soon after dicots germinate, the hypocotyl arches into a hook-like structure that protects the shoot apical meristem as the seedling grows through the soil. Once the seedling emerges from the ground and senses light, the hypocotyl straightens. The asymmetric growth that results in apical hook formation depends on an auxin gradient; auxin response maxima in the hypocotyl locally reduce cell elongation, resulting in hypocotyl bending. Members of the auxin influx carrier AUX/LIKE-AUX1 (LAX) and efflux carrier PIN-FORMED (PIN) families, which direct polar auxin transport and thus influence the distribution of auxin, have been implicated in apical hook formation (Vandenbussche et al., 2010). Trans-Golgi network (TGN)-localized ECHIDNA (ECH) mediates the secretion of AUX1 during apical hook formation (Boutte et al., 2013), whereas members of the GNOM and BIG ADP-ribosylation factor-guanine exchange factor (ARF-GEFs) families function in the post-Golgi trafficking of PIN1, with the former mediating its polar recycling (Geldner et al., 2003) and the latter its secretion. Evidence suggests that a Brefeldin-A (BFA)-resistant ARF-GEF also functions in AUX1 trafficking (Kleine-Vehn et al., 2006). Ethylene promotes apical hook formation by regulating auxin responses and distribution (Harper et al., 2000).
To tease apart the molecular mechanism underlying apical hook formation, Jonsson et al. (2017) examined the contribution of ECH and ARF-GEFs to this process in Arabidopsis thaliana seedlings. Using a series of mutants and pharmacological treatments, they showed that the BIG1–4 ARF-GEFs act redundantly to maintain the apical hook once it has formed. Treatment with the ethylene precursor aminocyclopropane-1-carboxylate (ACC), which normally exaggerates the apical hook, had no effect when BIG function was perturbed, suggesting that BIG ARF-GEFs function in ethylene-mediated hook development.
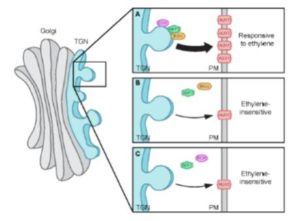 Given that apical hook development is insensitive to ethylene in both the big and ech mutants, and that both BIG and ECH proteins localize to the TGN (Boutte et al., 2013), the authors tested the possibility that these proteins function in a common pathway during apical hook development. The finding that hook developmental defects were more pronounced in the ech big2 big3 triple mutant than in ech or big2 big3 suggests that ECH and BIG proteins indeed act synergistically in apical hook development. By monitoring AUX1-YFP levels in transgenic lines in the wild-type and big3 background, the authors then showed that AUX-YFP levels were greatly reduced at the plasma membrane of the mutant, whereas transcript levels were only mildly reduced. Thus, BIG3—like ECH—mediates the secretion of AUX1 during apical hook formation. Further mutant studies using fluorescently-tagged ECH and AUX1 revealed that the colocalization of these proteins to the TGN is interdependent. Next, the authors examined the role of ARF1, which functions in vesicle formation and is the target ARF GTPase of BIG3 (Nielsen et al., 2006). They found that ARF1 localized to the TGN in a BIG- and ECH-dependent manner and that, like BIG and ECH, ARF1 functions in ethylene-mediated apical hook development and is required for the delivery of AUX1 to the plasma membrane.
Given that apical hook development is insensitive to ethylene in both the big and ech mutants, and that both BIG and ECH proteins localize to the TGN (Boutte et al., 2013), the authors tested the possibility that these proteins function in a common pathway during apical hook development. The finding that hook developmental defects were more pronounced in the ech big2 big3 triple mutant than in ech or big2 big3 suggests that ECH and BIG proteins indeed act synergistically in apical hook development. By monitoring AUX1-YFP levels in transgenic lines in the wild-type and big3 background, the authors then showed that AUX-YFP levels were greatly reduced at the plasma membrane of the mutant, whereas transcript levels were only mildly reduced. Thus, BIG3—like ECH—mediates the secretion of AUX1 during apical hook formation. Further mutant studies using fluorescently-tagged ECH and AUX1 revealed that the colocalization of these proteins to the TGN is interdependent. Next, the authors examined the role of ARF1, which functions in vesicle formation and is the target ARF GTPase of BIG3 (Nielsen et al., 2006). They found that ARF1 localized to the TGN in a BIG- and ECH-dependent manner and that, like BIG and ECH, ARF1 functions in ethylene-mediated apical hook development and is required for the delivery of AUX1 to the plasma membrane.
The authors weaved these findings into an elegant model (see figure), in which ECH and BIG recruit ARF1 to the TGN, where it mediates the vesicle formation needed for AUX1 delivery to the plasma membrane during ethylene-responsive apical hook development.
IN BRIEF by Kathleen L. Farquharson [email protected]
References
Boutte, Y., Jonsson, K., McFarlane, H.E., Johnson, E., Gendre, D., Swarup, R., Friml, J., Samuels, L., Robert, S., and Bhalerao, R.P. (2013). ECHIDNA-mediated post-Golgi trafficking of auxin carriers for differential cell elongation. PNAS 110, 16259–16264.
Geldner, N., Anders, N., Wolters, H., Keicher, J., Kornberger, W., Muller, P., Delbarre, A., Ueda, T., Nakano, A., and Jurgens, G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112, 219–230.
Harper, R.M., Stowe-Evans, E.L., Luesse, D.R., Muto, H., Tatematsu, K., Watahiki, M.K., Yamamoto, K., and Liscum, E. (2000). The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. The Plant Cell 12, 757–770.
Jonsson, K., Boutté, Y., Singh, R.K., Gendre, D., Bhalerao, R.P. (2017). Ethylene Regulates differential growth via BIG ARF-GEF-dependent post-Golgi secretory trafficking in Arabidopsis. The Plant Cell: tpc.00743.2016.
Kleine-Vehn, J., Dhonukshe, P., Swarup, R., Bennett, M., and Friml, J. (2006). Subcellular trafficking of the Arabidopsis auxin influx carrier AUX1 uses a novel pathway distinct from PIN1. The Plant Cell 18, 3171–3181.
Nielsen, M., Albrethsen, J., Larsen, F.H., and Skriver, K. (2006). The Arabidopsis ADP-ribosylation factor (ARF) and ARF-like (ARL) system and its regulation by BIG2, a large ARF-GEF. Plant Sci 171, 707–717.
Vandenbussche, F., Petrasek, J., Zadnikova, P., Hoyerova, K., Pesek, B., Raz, V., Swarup, R., Bennett, M., Zazimalova, E., Benkova, E., and Van Der Straeten, D. (2010). The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development 137, 597–606.


 Sabeeha Merchant is a Professor of Biochemistry and a member of the Molecular Biology Institute at the University of California, Los Angeles. She completed BS and PhD degrees at the University of Wisconsin-Madison and was a Post-doctoral Scholar at Harvard University. She has conducted seminal research on the role of metals in regulating the biosynthesis and assembly of metalloproteins in photosynthetic eukaryotes, and pioneered discoveries in the assembly of metalloenzymes and the regulated biogenesis of major complexes of the photosynthetic apparatus in green algae. At present, the Merchant group uses a combination of classical genetics, genomics and biochemistry to discover mechanisms of trace metal homeostasis in Chlamydomonas. Recent work involves exploiting next generation sequencing technology (RNA-Seq) to define the transcriptome of wild-type and mutant Chlamydomonas strains under various nutrient, environmental, and developmental conditions with the goal of identifying the pattern of expression of every gene in the genome, divining co-expressed genes and key cis-regulatory sequences, and using transcriptome information for genome annotation.
Sabeeha Merchant is a Professor of Biochemistry and a member of the Molecular Biology Institute at the University of California, Los Angeles. She completed BS and PhD degrees at the University of Wisconsin-Madison and was a Post-doctoral Scholar at Harvard University. She has conducted seminal research on the role of metals in regulating the biosynthesis and assembly of metalloproteins in photosynthetic eukaryotes, and pioneered discoveries in the assembly of metalloenzymes and the regulated biogenesis of major complexes of the photosynthetic apparatus in green algae. At present, the Merchant group uses a combination of classical genetics, genomics and biochemistry to discover mechanisms of trace metal homeostasis in Chlamydomonas. Recent work involves exploiting next generation sequencing technology (RNA-Seq) to define the transcriptome of wild-type and mutant Chlamydomonas strains under various nutrient, environmental, and developmental conditions with the goal of identifying the pattern of expression of every gene in the genome, divining co-expressed genes and key cis-regulatory sequences, and using transcriptome information for genome annotation. Areas of expertise
Areas of expertise
 Areas of expertise
Areas of expertise Areas of expertise
Areas of expertise Areas of expertise
Areas of expertise Areas of expertise
Areas of expertise Areas of expertise
Areas of expertise