Review: The evolution of calcium-based signalling in plants
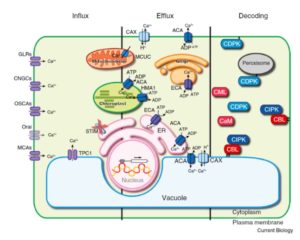 Calcium signalling is a ubiquitous process in plants and other organisms. Transporters at the plasma and vacuolar (tonoplast) membranes control entry and exit of calcium ion into the cytoplasm, and it is the cytoplasmic calcium level ([Ca2+]cyt) that affects cellular responses, as calcium-binding proteins alter their activities or binding properties in response to changing calcium levels. Edel et al. review the diversity and distribution of various components of the calcium-based signalling process in plants as compared to animals, and speculate as to the forces that have shaped it in plants, including photosynthesis. Curr. Biol. 10.1016/j.cub.2017.05.020
Calcium signalling is a ubiquitous process in plants and other organisms. Transporters at the plasma and vacuolar (tonoplast) membranes control entry and exit of calcium ion into the cytoplasm, and it is the cytoplasmic calcium level ([Ca2+]cyt) that affects cellular responses, as calcium-binding proteins alter their activities or binding properties in response to changing calcium levels. Edel et al. review the diversity and distribution of various components of the calcium-based signalling process in plants as compared to animals, and speculate as to the forces that have shaped it in plants, including photosynthesis. Curr. Biol. 10.1016/j.cub.2017.05.020
Review: Uncovering gene regulatory networks controlling plant cell differentiation ($)
 The transparency and simplicity of the Arabidopsis root apex has made it an excellent model through which to understand the processes that lead from undifferentiated meristem cells to mature root tissues. Drapek et al. review the gene regulatory networks (GRNs) that underlie the development of the endodermis in Arabidopsis root tips. They first summarize the roles of transcription factors including mobile transcription factors (those that move between cells) in the specification of the endodermal cells, and then they discuss the GRNs that control the differentiation of endodermal cells including the proper formation of the Casparian strip. They conclude by describing outstanding questions and new approaches. Trends Genetics 10.1016/j.tig.2017.05.002
The transparency and simplicity of the Arabidopsis root apex has made it an excellent model through which to understand the processes that lead from undifferentiated meristem cells to mature root tissues. Drapek et al. review the gene regulatory networks (GRNs) that underlie the development of the endodermis in Arabidopsis root tips. They first summarize the roles of transcription factors including mobile transcription factors (those that move between cells) in the specification of the endodermal cells, and then they discuss the GRNs that control the differentiation of endodermal cells including the proper formation of the Casparian strip. They conclude by describing outstanding questions and new approaches. Trends Genetics 10.1016/j.tig.2017.05.002
Update: Shining light on the function of NPH3/RPT2-like proteins in phototropin signalling
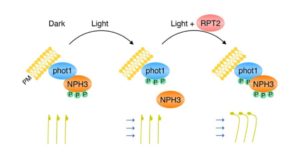 Blue light regulates diverse functions including phototropism and chloroplast movements. Christie et al. summarize recent updates in understanding the signalling pathway between light perception by phototropins and downstream responses. They describe the central roles for the 33 members (in Arabidopsis) of the NRL protein family, named for the founding members NON-PHOTOTROPIC HYPOCOTYL (NPH3) and ROOT PHOTOTROPISM 2 (RPT2), and the four members of the PHYTOCHROME KINASE SUBSRTATE (PKS) protein family. The authors also summarize the evolutionary origins of the blue-light signalling pathway in the green plant lineage and highlight ongoing questions including the need for biochemical characterization of many of these proteins. Plant Physiol. 10.1104/pp.17.00835
Blue light regulates diverse functions including phototropism and chloroplast movements. Christie et al. summarize recent updates in understanding the signalling pathway between light perception by phototropins and downstream responses. They describe the central roles for the 33 members (in Arabidopsis) of the NRL protein family, named for the founding members NON-PHOTOTROPIC HYPOCOTYL (NPH3) and ROOT PHOTOTROPISM 2 (RPT2), and the four members of the PHYTOCHROME KINASE SUBSRTATE (PKS) protein family. The authors also summarize the evolutionary origins of the blue-light signalling pathway in the green plant lineage and highlight ongoing questions including the need for biochemical characterization of many of these proteins. Plant Physiol. 10.1104/pp.17.00835
Review: Unlocking the potential of plant phenotyping data through integration and data-driven approaches
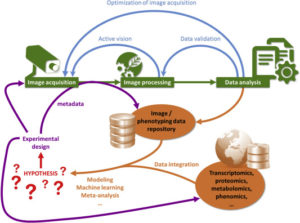 Coppens et al. describe the challenges and opportunities for developing integrated data accession and analysis systems for plant phenotyping. They observe that the lack of standardization for ontologies, data forms and structures and metadata collection needs to be resolved, both to increase the value of collected data and also for the application of machine learning. The article includes links to many useful repositories, datasets, projects and platforms. Curr. Opin. Sys. Biol. 10.1016/j.coisb.2017.07.002
Coppens et al. describe the challenges and opportunities for developing integrated data accession and analysis systems for plant phenotyping. They observe that the lack of standardization for ontologies, data forms and structures and metadata collection needs to be resolved, both to increase the value of collected data and also for the application of machine learning. The article includes links to many useful repositories, datasets, projects and platforms. Curr. Opin. Sys. Biol. 10.1016/j.coisb.2017.07.002
Single-molecule spectroscopy of LHCSR1 identifies two distinct states responsible for photosynthetic photoprotection
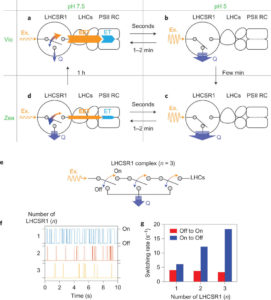 Light-harvesting complex stress-related (LHCSR) proteins catalyze excess-light energy dissipation in green algae and mosses. Kondo et al. used use single-molecule spectroscopy to uncover how LHCSR1 achieves this. The authors find the protein exists in two states, one activated by a change in pH and the other by carotenoid composition (specifically, the xanthophyll cycle). The authors describe how regulated conformational dynamics match LHCSR1 function to different conditions. Nature Chem. 10.1038/nchem.2818 (Here is a plain-language summary about this work).
Light-harvesting complex stress-related (LHCSR) proteins catalyze excess-light energy dissipation in green algae and mosses. Kondo et al. used use single-molecule spectroscopy to uncover how LHCSR1 achieves this. The authors find the protein exists in two states, one activated by a change in pH and the other by carotenoid composition (specifically, the xanthophyll cycle). The authors describe how regulated conformational dynamics match LHCSR1 function to different conditions. Nature Chem. 10.1038/nchem.2818 (Here is a plain-language summary about this work).
Rapid recovery gene downregulation during excess-light stress and recovery in Arabidopsis
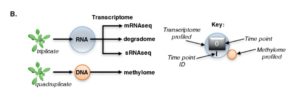 Previous exposure to stress can sometimes lead to enhanced stress tolerance through memory or priming, but the molecular mechanisms through which this occurs remain uncertain. Crisp et al. investigated how the transcriptome resets itself after high-light stress. They identified a subset of mRNAs that rapidly decay following the alleviation of stress, in an affect that they call Rapid Recovery Gene Downregulation (RRGD). The authors found little evidence for small RNAs or 5ʹ-3ʹ RNA decay contributing to rapid decay of mRNAs, but they do find evidence for ribosome stalling and co-translational decay having a role. They also found little evidence for the involvement of DNA methylation. Although the mechanism of the observed rapid gene downregulation remains unclear, the large datasets of mRNA dynamics during stress recovery should help to elucidate the factors that contribute to memory and priming. Plant Cell 10.1105/tpc.16.00828
Previous exposure to stress can sometimes lead to enhanced stress tolerance through memory or priming, but the molecular mechanisms through which this occurs remain uncertain. Crisp et al. investigated how the transcriptome resets itself after high-light stress. They identified a subset of mRNAs that rapidly decay following the alleviation of stress, in an affect that they call Rapid Recovery Gene Downregulation (RRGD). The authors found little evidence for small RNAs or 5ʹ-3ʹ RNA decay contributing to rapid decay of mRNAs, but they do find evidence for ribosome stalling and co-translational decay having a role. They also found little evidence for the involvement of DNA methylation. Although the mechanism of the observed rapid gene downregulation remains unclear, the large datasets of mRNA dynamics during stress recovery should help to elucidate the factors that contribute to memory and priming. Plant Cell 10.1105/tpc.16.00828
Novel stress-inducible antisense RNAs of protein-coding loci are synthesized by RNA-dependent RNA polymerase
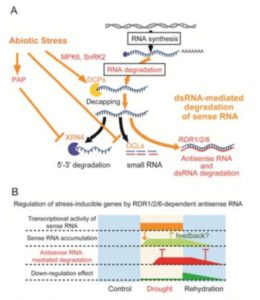 Previous work showed that many long non-coding RNAs are produced in response to abiotic stress, and that these often show considerable overlap with protein-coding genes. Matsui et al. investigated two of these, fAsRD29A1 and fAsRD29A2, corresponding to (A1) a full-length antisense RNA of the drought-inducible gene RD29A and (A2) one corresponding to the mRNA with the intron spliced out. They examined production of these AS RNAs in several different mutant backgrounds to identify their mechanisms of production; key findings include that endogenous uncapped sense RNA serves as a template for their synthesis, and that although RNA-dependent RNA Polymerases (RDR 1/2/6) are required for their production, this occurs independently of siRNA production. The authors also find that the AS RNAs contribute to turnover of the cognate mRNAs. They summarize, “These data reveal a novel mechanism of RNA regulation during abiotic stress response that involves complex RNA degradation pathways.” Plant Physiol. 10.1104/pp.17.00787
Previous work showed that many long non-coding RNAs are produced in response to abiotic stress, and that these often show considerable overlap with protein-coding genes. Matsui et al. investigated two of these, fAsRD29A1 and fAsRD29A2, corresponding to (A1) a full-length antisense RNA of the drought-inducible gene RD29A and (A2) one corresponding to the mRNA with the intron spliced out. They examined production of these AS RNAs in several different mutant backgrounds to identify their mechanisms of production; key findings include that endogenous uncapped sense RNA serves as a template for their synthesis, and that although RNA-dependent RNA Polymerases (RDR 1/2/6) are required for their production, this occurs independently of siRNA production. The authors also find that the AS RNAs contribute to turnover of the cognate mRNAs. They summarize, “These data reveal a novel mechanism of RNA regulation during abiotic stress response that involves complex RNA degradation pathways.” Plant Physiol. 10.1104/pp.17.00787
Maize YABBY genes drooping leaf1 and drooping leaf2 regulate plant architecture
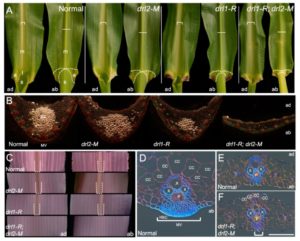 Strable et al. describe the characterization of two loci that affect leaf morphology and plant architecture. The leaf insertion angle in droopingleaf1 (drl1-R) mutants is larger than that in wild-type, making the leaves appear droopy, an effect that is associated with a reduced midvein. This effect is enhanced in the presence of the drl2-M mutation. Both the DRL genes encode YABBY transcription factors and are located in quantitative trait loci (QTL) regions previously identified as affecting leaf angle. The authors observe that these and related loci are candidates for optimizing leaf-angle effects on light capture for photosynthesis. Plant Physiol. 10.1105/tpc.16.00477
Strable et al. describe the characterization of two loci that affect leaf morphology and plant architecture. The leaf insertion angle in droopingleaf1 (drl1-R) mutants is larger than that in wild-type, making the leaves appear droopy, an effect that is associated with a reduced midvein. This effect is enhanced in the presence of the drl2-M mutation. Both the DRL genes encode YABBY transcription factors and are located in quantitative trait loci (QTL) regions previously identified as affecting leaf angle. The authors observe that these and related loci are candidates for optimizing leaf-angle effects on light capture for photosynthesis. Plant Physiol. 10.1105/tpc.16.00477
A statistical description of plant shoot architecture ($)
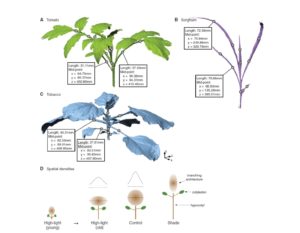 Conn et al. analysed nearly 600 three-dimensional scans of three plant species (tomato, tobacco and sorghum) from different developmental stages and environments, and identified three underlying principles. The first they call separability, which indicates that variation in one dimension can occur without affecting the other dimensions. The second they call self-similarity, which indicates that the shapes are fundamentally similar and differ only by stretching and compression. The third is that all plant architectures follow a single form, that of a 3D Gaussian truncated at roughly two standard deviations (SDs) from the center of mass, so that “only four parameters are needed to specify statistically any architecture’s size and shape: its total branch length and the SDs of the Gaussian in the three orthogonal directions.” They finally compare these architectural principles to those of other naturally occurring structures. Curr. Biol. 10.1016/j.cub.2017.06.009
Conn et al. analysed nearly 600 three-dimensional scans of three plant species (tomato, tobacco and sorghum) from different developmental stages and environments, and identified three underlying principles. The first they call separability, which indicates that variation in one dimension can occur without affecting the other dimensions. The second they call self-similarity, which indicates that the shapes are fundamentally similar and differ only by stretching and compression. The third is that all plant architectures follow a single form, that of a 3D Gaussian truncated at roughly two standard deviations (SDs) from the center of mass, so that “only four parameters are needed to specify statistically any architecture’s size and shape: its total branch length and the SDs of the Gaussian in the three orthogonal directions.” They finally compare these architectural principles to those of other naturally occurring structures. Curr. Biol. 10.1016/j.cub.2017.06.009
Evolution of the wheat blast fungus through functional losses in a host specificity determinant ($)
 We describe plant-pathogen interactions as an “arms race” whereby each new weapon deployed by the pathogen is countered by enhanced host defences. Inoue et al. reveal the molecular footprint of the arms race between a blast fungus and its hosts, in particular how a fungal pathogen of rice, oat and ryegrass (Pyricularia oryzae) evolved to also infect wheat. The authors found that the origins of this host jump correlate with the widespread cultivation of a wheat variety that fails to recognizes certain fungal proteins, which allowed the fungal population to increase and adapt to its new host. As the accompanying summary by Maekawa and Schulze-Lefert concludes, “The dynamics of P. oryzae host range alterations is likely influenced by plant domestication and agricultural practices favoring cultivars with homogenous genetic makeup that carry only a fraction of the resistance gene repertoire present in their wild relatives” – an important reminder that genetically homogenous populations are fertile breeding grounds for pathogens. Science 10.1126/science.aam9654
We describe plant-pathogen interactions as an “arms race” whereby each new weapon deployed by the pathogen is countered by enhanced host defences. Inoue et al. reveal the molecular footprint of the arms race between a blast fungus and its hosts, in particular how a fungal pathogen of rice, oat and ryegrass (Pyricularia oryzae) evolved to also infect wheat. The authors found that the origins of this host jump correlate with the widespread cultivation of a wheat variety that fails to recognizes certain fungal proteins, which allowed the fungal population to increase and adapt to its new host. As the accompanying summary by Maekawa and Schulze-Lefert concludes, “The dynamics of P. oryzae host range alterations is likely influenced by plant domestication and agricultural practices favoring cultivars with homogenous genetic makeup that carry only a fraction of the resistance gene repertoire present in their wild relatives” – an important reminder that genetically homogenous populations are fertile breeding grounds for pathogens. Science 10.1126/science.aam9654
CRISPR–Cas encoding of a digital movie into genomes of a population of living bacteria ($)
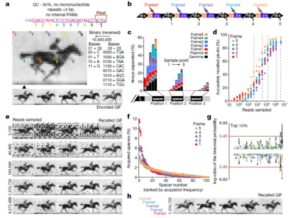 This is a fun and interesting look at how a population of bacteria can serve as a repository for digital data. Shipman et al. generated a code by which graphical information (pixels) could be translated to nucleotide sequences. They then introduced DNA encoding the pixel information into bacterial cells using CRISPR-Cas. Because the amount of data in the encoded images is large, they put a pool of DNA sequences into a population of cells. Sequencing the engineered locus from the pool returned all of the data, which was than decoded into an image strikingly like the starting image. This study demonstrates the potential of engineered bacteria as data storage devices. It is interesting to consider the implications of this work, some of which are described by Gina Kolata in a New York Times article about this work https://www.nytimes.com/2017/07/12/science/film-clip-stored-in-dna.html. Nature 10.1038/nature23017
This is a fun and interesting look at how a population of bacteria can serve as a repository for digital data. Shipman et al. generated a code by which graphical information (pixels) could be translated to nucleotide sequences. They then introduced DNA encoding the pixel information into bacterial cells using CRISPR-Cas. Because the amount of data in the encoded images is large, they put a pool of DNA sequences into a population of cells. Sequencing the engineered locus from the pool returned all of the data, which was than decoded into an image strikingly like the starting image. This study demonstrates the potential of engineered bacteria as data storage devices. It is interesting to consider the implications of this work, some of which are described by Gina Kolata in a New York Times article about this work https://www.nytimes.com/2017/07/12/science/film-clip-stored-in-dna.html. Nature 10.1038/nature23017




 The transparency and simplicity of the Arabidopsis root apex has made it an excellent model through which to understand the processes that lead from undifferentiated meristem cells to mature root tissues. Drapek et al. review the gene regulatory networks (GRNs) that underlie the development of the endodermis in Arabidopsis root tips. They first summarize the roles of transcription factors including mobile transcription factors (those that move between cells) in the specification of the endodermal cells, and then they discuss the GRNs that control the differentiation of endodermal cells including the proper formation of the Casparian strip. They conclude by describing outstanding questions and new approaches. Trends Genetics
The transparency and simplicity of the Arabidopsis root apex has made it an excellent model through which to understand the processes that lead from undifferentiated meristem cells to mature root tissues. Drapek et al. review the gene regulatory networks (GRNs) that underlie the development of the endodermis in Arabidopsis root tips. They first summarize the roles of transcription factors including mobile transcription factors (those that move between cells) in the specification of the endodermal cells, and then they discuss the GRNs that control the differentiation of endodermal cells including the proper formation of the Casparian strip. They conclude by describing outstanding questions and new approaches. Trends Genetics  Blue light regulates diverse functions including phototropism and chloroplast movements. Christie et al. summarize recent updates in understanding the signalling pathway between light perception by phototropins and downstream responses. They describe the central roles for the 33 members (in Arabidopsis) of the NRL protein family, named for the founding members NON-PHOTOTROPIC HYPOCOTYL (NPH3) and ROOT PHOTOTROPISM 2 (RPT2), and the four members of the PHYTOCHROME KINASE SUBSRTATE (PKS) protein family. The authors also summarize the evolutionary origins of the blue-light signalling pathway in the green plant lineage and highlight ongoing questions including the need for biochemical characterization of many of these proteins. Plant Physiol.
Blue light regulates diverse functions including phototropism and chloroplast movements. Christie et al. summarize recent updates in understanding the signalling pathway between light perception by phototropins and downstream responses. They describe the central roles for the 33 members (in Arabidopsis) of the NRL protein family, named for the founding members NON-PHOTOTROPIC HYPOCOTYL (NPH3) and ROOT PHOTOTROPISM 2 (RPT2), and the four members of the PHYTOCHROME KINASE SUBSRTATE (PKS) protein family. The authors also summarize the evolutionary origins of the blue-light signalling pathway in the green plant lineage and highlight ongoing questions including the need for biochemical characterization of many of these proteins. Plant Physiol.  Calcium signalling is a ubiquitous process in plants and other organisms. Transporters at the plasma and vacuolar (tonoplast) membranes control entry and exit of calcium ion into the cytoplasm, and it is the cytoplasmic calcium level ([Ca2+]cyt) that affects cellular responses, as calcium-binding proteins alter their activities or binding properties in response to changing calcium levels. Edel et al. review the diversity and distribution of various components of the calcium-based signalling process in plants as compared to animals, and speculate as to the forces that have shaped it in plants, including photosynthesis. Curr. Biol.
Calcium signalling is a ubiquitous process in plants and other organisms. Transporters at the plasma and vacuolar (tonoplast) membranes control entry and exit of calcium ion into the cytoplasm, and it is the cytoplasmic calcium level ([Ca2+]cyt) that affects cellular responses, as calcium-binding proteins alter their activities or binding properties in response to changing calcium levels. Edel et al. review the diversity and distribution of various components of the calcium-based signalling process in plants as compared to animals, and speculate as to the forces that have shaped it in plants, including photosynthesis. Curr. Biol.  Coppens et al. describe the challenges and opportunities for developing integrated data accession and analysis systems for plant phenotyping. They observe that the lack of standardization for ontologies, data forms and structures and metadata collection needs to be resolved, both to increase the value of collected data and also for the application of machine learning. The article includes links to many useful repositories, datasets, projects and platforms. Curr. Opin. Sys. Biol.
Coppens et al. describe the challenges and opportunities for developing integrated data accession and analysis systems for plant phenotyping. They observe that the lack of standardization for ontologies, data forms and structures and metadata collection needs to be resolved, both to increase the value of collected data and also for the application of machine learning. The article includes links to many useful repositories, datasets, projects and platforms. Curr. Opin. Sys. Biol.  Light-harvesting complex stress-related (LHCSR) proteins catalyze excess-light energy dissipation in green algae and mosses. Kondo et al. used use single-molecule spectroscopy to uncover how LHCSR1 achieves this. The authors find the protein exists in two states, one activated by a change in pH and the other by carotenoid composition (specifically, the xanthophyll cycle). The authors describe how regulated conformational dynamics match LHCSR1 function to different conditions. Nature Chem.
Light-harvesting complex stress-related (LHCSR) proteins catalyze excess-light energy dissipation in green algae and mosses. Kondo et al. used use single-molecule spectroscopy to uncover how LHCSR1 achieves this. The authors find the protein exists in two states, one activated by a change in pH and the other by carotenoid composition (specifically, the xanthophyll cycle). The authors describe how regulated conformational dynamics match LHCSR1 function to different conditions. Nature Chem.  Previous exposure to stress can sometimes lead to enhanced stress tolerance through memory or priming, but the molecular mechanisms through which this occurs remain uncertain. Crisp et al. investigated how the transcriptome resets itself after high-light stress. They identified a subset of mRNAs that rapidly decay following the alleviation of stress, in an affect that they call Rapid Recovery Gene Downregulation (RRGD). The authors found little evidence for small RNAs or 5ʹ-3ʹ RNA decay contributing to rapid decay of mRNAs, but they do find evidence for ribosome stalling and co-translational decay having a role. They also found little evidence for the involvement of DNA methylation. Although the mechanism of the observed rapid gene downregulation remains unclear, the large datasets of mRNA dynamics during stress recovery should help to elucidate the factors that contribute to memory and priming. Plant Cell
Previous exposure to stress can sometimes lead to enhanced stress tolerance through memory or priming, but the molecular mechanisms through which this occurs remain uncertain. Crisp et al. investigated how the transcriptome resets itself after high-light stress. They identified a subset of mRNAs that rapidly decay following the alleviation of stress, in an affect that they call Rapid Recovery Gene Downregulation (RRGD). The authors found little evidence for small RNAs or 5ʹ-3ʹ RNA decay contributing to rapid decay of mRNAs, but they do find evidence for ribosome stalling and co-translational decay having a role. They also found little evidence for the involvement of DNA methylation. Although the mechanism of the observed rapid gene downregulation remains unclear, the large datasets of mRNA dynamics during stress recovery should help to elucidate the factors that contribute to memory and priming. Plant Cell  Previous work showed that many long non-coding RNAs are produced in response to abiotic stress, and that these often show considerable overlap with protein-coding genes. Matsui et al. investigated two of these, fAsRD29A1 and fAsRD29A2, corresponding to (A1) a full-length antisense RNA of the drought-inducible gene RD29A and (A2) one corresponding to the mRNA with the intron spliced out. They examined production of these AS RNAs in several different mutant backgrounds to identify their mechanisms of production; key findings include that endogenous uncapped sense RNA serves as a template for their synthesis, and that although RNA-dependent RNA Polymerases (RDR 1/2/6) are required for their production, this occurs independently of siRNA production. The authors also find that the AS RNAs contribute to turnover of the cognate mRNAs. They summarize, “These data reveal a novel mechanism of RNA regulation during abiotic stress response that involves complex RNA degradation pathways.” Plant Physiol.
Previous work showed that many long non-coding RNAs are produced in response to abiotic stress, and that these often show considerable overlap with protein-coding genes. Matsui et al. investigated two of these, fAsRD29A1 and fAsRD29A2, corresponding to (A1) a full-length antisense RNA of the drought-inducible gene RD29A and (A2) one corresponding to the mRNA with the intron spliced out. They examined production of these AS RNAs in several different mutant backgrounds to identify their mechanisms of production; key findings include that endogenous uncapped sense RNA serves as a template for their synthesis, and that although RNA-dependent RNA Polymerases (RDR 1/2/6) are required for their production, this occurs independently of siRNA production. The authors also find that the AS RNAs contribute to turnover of the cognate mRNAs. They summarize, “These data reveal a novel mechanism of RNA regulation during abiotic stress response that involves complex RNA degradation pathways.” Plant Physiol.  Strable et al. describe the characterization of two loci that affect leaf morphology and plant architecture. The leaf insertion angle in droopingleaf1 (drl1-R) mutants is larger than that in wild-type, making the leaves appear droopy, an effect that is associated with a reduced midvein. This effect is enhanced in the presence of the drl2-M mutation. Both the DRL genes encode YABBY transcription factors and are located in quantitative trait loci (QTL) regions previously identified as affecting leaf angle. The authors observe that these and related loci are candidates for optimizing leaf-angle effects on light capture for photosynthesis. Plant Physiol.
Strable et al. describe the characterization of two loci that affect leaf morphology and plant architecture. The leaf insertion angle in droopingleaf1 (drl1-R) mutants is larger than that in wild-type, making the leaves appear droopy, an effect that is associated with a reduced midvein. This effect is enhanced in the presence of the drl2-M mutation. Both the DRL genes encode YABBY transcription factors and are located in quantitative trait loci (QTL) regions previously identified as affecting leaf angle. The authors observe that these and related loci are candidates for optimizing leaf-angle effects on light capture for photosynthesis. Plant Physiol.  Conn et al. analysed nearly 600 three-dimensional scans of three plant species (tomato, tobacco and sorghum) from different developmental stages and environments, and identified three underlying principles. The first they call separability, which indicates that variation in one dimension can occur without affecting the other dimensions. The second they call self-similarity, which indicates that the shapes are fundamentally similar and differ only by stretching and compression. The third is that all plant architectures follow a single form, that of a 3D Gaussian truncated at roughly two standard deviations (SDs) from the center of mass, so that “only four parameters are needed to specify statistically any architecture’s size and shape: its total branch length and the SDs of the Gaussian in the three orthogonal directions.” They finally compare these architectural principles to those of other naturally occurring structures. Curr. Biol.
Conn et al. analysed nearly 600 three-dimensional scans of three plant species (tomato, tobacco and sorghum) from different developmental stages and environments, and identified three underlying principles. The first they call separability, which indicates that variation in one dimension can occur without affecting the other dimensions. The second they call self-similarity, which indicates that the shapes are fundamentally similar and differ only by stretching and compression. The third is that all plant architectures follow a single form, that of a 3D Gaussian truncated at roughly two standard deviations (SDs) from the center of mass, so that “only four parameters are needed to specify statistically any architecture’s size and shape: its total branch length and the SDs of the Gaussian in the three orthogonal directions.” They finally compare these architectural principles to those of other naturally occurring structures. Curr. Biol.  We describe plant-pathogen interactions as an “arms race” whereby each new weapon deployed by the pathogen is countered by enhanced host defences. Inoue et al. reveal the molecular footprint of the arms race between a blast fungus and its hosts, in particular how a fungal pathogen of rice, oat and ryegrass (Pyricularia oryzae) evolved to also infect wheat. The authors found that the origins of this host jump correlate with the widespread cultivation of a wheat variety that fails to recognizes certain fungal proteins, which allowed the fungal population to increase and adapt to its new host. As the accompanying summary by Maekawa and Schulze-Lefert concludes, “The dynamics of P. oryzae host range alterations is likely influenced by plant domestication and agricultural practices favoring cultivars with homogenous genetic makeup that carry only a fraction of the resistance gene repertoire present in their wild relatives” – an important reminder that genetically homogenous populations are fertile breeding grounds for pathogens. Science
We describe plant-pathogen interactions as an “arms race” whereby each new weapon deployed by the pathogen is countered by enhanced host defences. Inoue et al. reveal the molecular footprint of the arms race between a blast fungus and its hosts, in particular how a fungal pathogen of rice, oat and ryegrass (Pyricularia oryzae) evolved to also infect wheat. The authors found that the origins of this host jump correlate with the widespread cultivation of a wheat variety that fails to recognizes certain fungal proteins, which allowed the fungal population to increase and adapt to its new host. As the accompanying summary by Maekawa and Schulze-Lefert concludes, “The dynamics of P. oryzae host range alterations is likely influenced by plant domestication and agricultural practices favoring cultivars with homogenous genetic makeup that carry only a fraction of the resistance gene repertoire present in their wild relatives” – an important reminder that genetically homogenous populations are fertile breeding grounds for pathogens. Science  This is a fun and interesting look at how a population of bacteria can serve as a repository for digital data. Shipman et al. generated a code by which graphical information (pixels) could be translated to nucleotide sequences. They then introduced DNA encoding the pixel information into bacterial cells using CRISPR-Cas. Because the amount of data in the encoded images is large, they put a pool of DNA sequences into a population of cells. Sequencing the engineered locus from the pool returned all of the data, which was than decoded into an image strikingly like the starting image. This study demonstrates the potential of engineered bacteria as data storage devices. It is interesting to consider the implications of this work, some of which are described by Gina Kolata in a New York Times article about this work
This is a fun and interesting look at how a population of bacteria can serve as a repository for digital data. Shipman et al. generated a code by which graphical information (pixels) could be translated to nucleotide sequences. They then introduced DNA encoding the pixel information into bacterial cells using CRISPR-Cas. Because the amount of data in the encoded images is large, they put a pool of DNA sequences into a population of cells. Sequencing the engineered locus from the pool returned all of the data, which was than decoded into an image strikingly like the starting image. This study demonstrates the potential of engineered bacteria as data storage devices. It is interesting to consider the implications of this work, some of which are described by Gina Kolata in a New York Times article about this work