Opinion: We aren’t good at picking candidate genes, and it’s slowing us down (COPB)
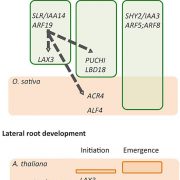
 Recent advances have facilitated the generation of huge phenotypic datasets from plant populations. However, the means to inexpensively organise such datasets to unequivocally determine causal genes has evaded researchers. Here, Baxter discusses how human bias when selecting candidate genes is compromising research efficiency. When utilising breeding populations for association mapping, a genomic region correlated to a trait is often identified. Subsequently, genes localized in the region are compared to databases to reduce the number of candidates into a manageable list. This process requires human judgement when selecting what databases to use and which genes to follow up on, introducing bias. For example, we tend to focus on genes previously documented, limiting progress in characterising genes. The first step towards overcoming such biases is to recognise their existence and consciously work against them. Next, the author suggests potential solutions to shorten candidate lists without the need for human judgement, such as employing transcriptomics, proteomics, epigenetics and phylogenetics. (Summary by Caroline Dowling) Curr. Opin. Plant Biol. 10.1016/j.pbi.2020.01.006
Recent advances have facilitated the generation of huge phenotypic datasets from plant populations. However, the means to inexpensively organise such datasets to unequivocally determine causal genes has evaded researchers. Here, Baxter discusses how human bias when selecting candidate genes is compromising research efficiency. When utilising breeding populations for association mapping, a genomic region correlated to a trait is often identified. Subsequently, genes localized in the region are compared to databases to reduce the number of candidates into a manageable list. This process requires human judgement when selecting what databases to use and which genes to follow up on, introducing bias. For example, we tend to focus on genes previously documented, limiting progress in characterising genes. The first step towards overcoming such biases is to recognise their existence and consciously work against them. Next, the author suggests potential solutions to shorten candidate lists without the need for human judgement, such as employing transcriptomics, proteomics, epigenetics and phylogenetics. (Summary by Caroline Dowling) Curr. Opin. Plant Biol. 10.1016/j.pbi.2020.01.006
[altmetric doi=”10.1016/j.pbi.2020.01.006″ details=”right” float=”right”]