New Insights into the Molecular Biology of Plant Circadian Rhythms
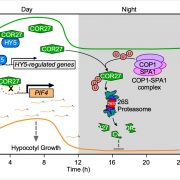
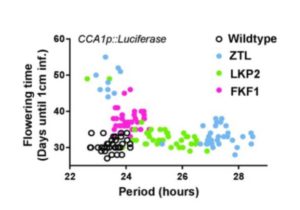 The circadian clock is an endogenous timekeeper that synchronizes essential biological processes with the outside world. Eukaryotic clocks rely on the ubiquitin proteasome system to target core clock factors for degradation. Altering clock protein degradation can change the period length of the clock. The ubiquitin proteasome system is an enzymatic pathway that mediates the covalent attachment of substrate proteins to ubiquitin, a step that targets them to the proteasome for destruction. All eukaryotic circadian clocks employ SKP1/CULLIN/F-BOX-type E3 ligases to mediate ubiquitylation of clock factors. In plants, the partially redundant F-box-type E3 ubiquitin ligases ZEITLUPE (ZTL), FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (FKF1), and LOV KELCH PROTEIN 2 (LKP2) regulate clock period and couple the clock to photoperiodic flowering in response to end-of-day light conditions. To better understand the functions of these three E3 ubiquitin ligases, Lee et al. (10.1104/pp.18.00331) expressed decoy ZTL, FKF1, and LKP2 proteins, lacking F-box domains, in Arabidopsis. These decoys associate with target proteins but are unable to ubiquitylate their targets, thereby inhibiting the ubiquitylation of target proteins and allowing for the study of the ubiquitylation-independent- and dependent- functions of ZTL, FKF1, and LKP2. The decoy E3 ligases also trap substrate interactions, allowing immunoprecipitation-mass spectrometry to be used to identify interacting partners. Much of the authors’ focus concerns the transcription factor CCA1 HIKING EXPEDITION (CHE), a critical clock regulator They show that ZTL interacts directly with CHE and mediates CHE ubiquitylation. They also demonstrate that CHE protein is degraded in the dark, and degradation is reduced in a ztl mutant plant, observations consistent with CHE being a ZTL target protein. This work demonstrates an effective strategy for unravelling the complex genetic redundancy of E3 ubiquitin ligases.
The circadian clock is an endogenous timekeeper that synchronizes essential biological processes with the outside world. Eukaryotic clocks rely on the ubiquitin proteasome system to target core clock factors for degradation. Altering clock protein degradation can change the period length of the clock. The ubiquitin proteasome system is an enzymatic pathway that mediates the covalent attachment of substrate proteins to ubiquitin, a step that targets them to the proteasome for destruction. All eukaryotic circadian clocks employ SKP1/CULLIN/F-BOX-type E3 ligases to mediate ubiquitylation of clock factors. In plants, the partially redundant F-box-type E3 ubiquitin ligases ZEITLUPE (ZTL), FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (FKF1), and LOV KELCH PROTEIN 2 (LKP2) regulate clock period and couple the clock to photoperiodic flowering in response to end-of-day light conditions. To better understand the functions of these three E3 ubiquitin ligases, Lee et al. (10.1104/pp.18.00331) expressed decoy ZTL, FKF1, and LKP2 proteins, lacking F-box domains, in Arabidopsis. These decoys associate with target proteins but are unable to ubiquitylate their targets, thereby inhibiting the ubiquitylation of target proteins and allowing for the study of the ubiquitylation-independent- and dependent- functions of ZTL, FKF1, and LKP2. The decoy E3 ligases also trap substrate interactions, allowing immunoprecipitation-mass spectrometry to be used to identify interacting partners. Much of the authors’ focus concerns the transcription factor CCA1 HIKING EXPEDITION (CHE), a critical clock regulator They show that ZTL interacts directly with CHE and mediates CHE ubiquitylation. They also demonstrate that CHE protein is degraded in the dark, and degradation is reduced in a ztl mutant plant, observations consistent with CHE being a ZTL target protein. This work demonstrates an effective strategy for unravelling the complex genetic redundancy of E3 ubiquitin ligases.