New Insights into Carboxysomes
 Despite its essential role in photosynthetic carbon fixation, ribulose 1,5 bisphosphate carboxylase/oxygenase (Rubisco) is a relatively inefficient enzyme, due in part to its inability to discriminate between CO2 and O2 as substrates. To suppress the oxygenase reaction and enhance the carboxylase activity of Rubisco, cyanobacteria have specialized bacterial microcompartments (BMCs) called carboxysomes that are central components of these organisms’ CO2-concentrating mechanisms (CCMs). Understanding the molecular mechanism underlying Rubisco assembly and carboxysome biogenesis may prove useful for engineering functional CO2-fixing complexes in heterogeneous organisms, especially plants, with the aim of boosting photosynthesis and agricultural productivity. There are two types of carboxysomes, distinguished by the form of Rubisco they encapsulate: Form IA in α-carboxysomes and Form IB in β-carboxysomes. In β-carboxysomes, form IB Rubisco and carbonic anhydrase are densely packed into an ordered matrix with internal linker proteins to form the enzyme core, which is encapsulated by a proteinaceous shell. The shell acts as a selective barrier that is permeable to bicarbonate and ribulose-1,5-bisphosphate, the substrates of Rubisco. The shells of carboxysomes consist of three protein types, each named after the oligomer they form: BMC-H (hexamer), BMC-P (pentamer), and BMC-T (trimer). These three protein types form cyclic homooligomers with pores at the center of symmetry that enable metabolite transport across the shell. Carboxysome shells contain multiple BMC-H paralogs, each with distinctly conserved residues surrounding the pore, which are assumed to be associated with specific metabolites. Sommer et al. (10.1104/pp.18.01190) have examined the regulation of β-carboxysome shell composition by investigating the BMC-H-encoding genes ccmK3 and ccmK4. A recent comprehensive survey of carboxysome genes in β-cyanobacteria showed that ccmK3 and ccmK4 genes always co-occur; although functionally linked, the two proteins are functionally linked. The present study provides evidence that CcmK3 does not form homohexamers, but rather that CcmK3 and CcmK4 form a heterohexameric complex with a 1:2 stoichiometry that can further form dodecamers under certain conditions. The authors suggest that heterohexamer formation could provide a means of fine-tuning β-carboxysome shell permeability. Moreover, the observed formation of dodecamers in solution suggests that carboxysome shell permeability may be dynamically attenuated by “capping” hexamers with a second hexamer. Heterohexamer formation and capping could provide a rapid and reversible means to alter metabolite flux across the shell in response to environmental/growth conditions.
Despite its essential role in photosynthetic carbon fixation, ribulose 1,5 bisphosphate carboxylase/oxygenase (Rubisco) is a relatively inefficient enzyme, due in part to its inability to discriminate between CO2 and O2 as substrates. To suppress the oxygenase reaction and enhance the carboxylase activity of Rubisco, cyanobacteria have specialized bacterial microcompartments (BMCs) called carboxysomes that are central components of these organisms’ CO2-concentrating mechanisms (CCMs). Understanding the molecular mechanism underlying Rubisco assembly and carboxysome biogenesis may prove useful for engineering functional CO2-fixing complexes in heterogeneous organisms, especially plants, with the aim of boosting photosynthesis and agricultural productivity. There are two types of carboxysomes, distinguished by the form of Rubisco they encapsulate: Form IA in α-carboxysomes and Form IB in β-carboxysomes. In β-carboxysomes, form IB Rubisco and carbonic anhydrase are densely packed into an ordered matrix with internal linker proteins to form the enzyme core, which is encapsulated by a proteinaceous shell. The shell acts as a selective barrier that is permeable to bicarbonate and ribulose-1,5-bisphosphate, the substrates of Rubisco. The shells of carboxysomes consist of three protein types, each named after the oligomer they form: BMC-H (hexamer), BMC-P (pentamer), and BMC-T (trimer). These three protein types form cyclic homooligomers with pores at the center of symmetry that enable metabolite transport across the shell. Carboxysome shells contain multiple BMC-H paralogs, each with distinctly conserved residues surrounding the pore, which are assumed to be associated with specific metabolites. Sommer et al. (10.1104/pp.18.01190) have examined the regulation of β-carboxysome shell composition by investigating the BMC-H-encoding genes ccmK3 and ccmK4. A recent comprehensive survey of carboxysome genes in β-cyanobacteria showed that ccmK3 and ccmK4 genes always co-occur; although functionally linked, the two proteins are functionally linked. The present study provides evidence that CcmK3 does not form homohexamers, but rather that CcmK3 and CcmK4 form a heterohexameric complex with a 1:2 stoichiometry that can further form dodecamers under certain conditions. The authors suggest that heterohexamer formation could provide a means of fine-tuning β-carboxysome shell permeability. Moreover, the observed formation of dodecamers in solution suggests that carboxysome shell permeability may be dynamically attenuated by “capping” hexamers with a second hexamer. Heterohexamer formation and capping could provide a rapid and reversible means to alter metabolite flux across the shell in response to environmental/growth conditions.
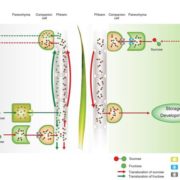
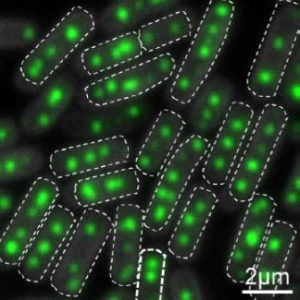
A second contribution in this issue relating to carboxysomes, concerns a chaperone protein RbcX that is essential for proper protein folding and Rubisco assembly. Huang et al. (10.1104/pp.18.01217) have investigated the in vivo function of RbcX in Synechococcus using molecular, biochemical, and live-cell fluorescence imaging approaches. Their results show that genetic deletion of the rbcX gene affects Rubisco abundance, as well as carboxysome formation and spatial distribution. Moreover, RbcX appears to be a component of the carboxysome and shows a dynamic interaction with Rubisco enzymes.