It takes two to be you: promoter motif pairs keep immune responses within cell identity boundaries
Being a multicellular organism is not an easy task. Proper functioning of individual cell types with different functions requires coordination of gene expression to determine cell identity, but also regulatory mechanisms to respond to environmental cues. The root may appear simple at first sight, but it is built with 15 cell types, each having an individual transcriptional profile, facilitating their functional specialization (Brady et al., 2007).
Root developmental processes are tightly interconnected with responses to the environment (Iyer-Pascuzzi et al., 2011). Several abiotic stresses have been shown to elicit cell type-specific responses. At the same time, root cell identity is maintained during stress and several cell identity regulators play an important role in stress responses (Iyer-Pascuzzi et al., 2011). Similarly, biotic stress responses show cell type‑specificity (Walker et al., 2017), yet it remains unknown how the root immunity and cell identity regulatory networks connect.
 To tackle this question, Rich-Griffin et al. (2020) investigated cell type-specific regulatory patterns activated by biotic stress. The authors treated Arabidopsis roots with elicitors of Pattern Triggered Immunity (PTI) signaling pathways leading to resistance to pathogens (Bigeard et al., 2015). With fluorescent-activated cell sorting, the authors profiled the transcriptomes of three root cell types—the two outermost root layers, epidermis and cortex, and the cell layer surrounding the root vasculature, the pericycle. More than half of the transcripts affected by PTI were expressed specifically in one out of three cell types. Moreover, expression of only a small proportion of the cell identity genes was changed by the immunity response. Together, these data suggest that despite strong cell type specificity in its response, PTI does not affect cell identity networks.
To tackle this question, Rich-Griffin et al. (2020) investigated cell type-specific regulatory patterns activated by biotic stress. The authors treated Arabidopsis roots with elicitors of Pattern Triggered Immunity (PTI) signaling pathways leading to resistance to pathogens (Bigeard et al., 2015). With fluorescent-activated cell sorting, the authors profiled the transcriptomes of three root cell types—the two outermost root layers, epidermis and cortex, and the cell layer surrounding the root vasculature, the pericycle. More than half of the transcripts affected by PTI were expressed specifically in one out of three cell types. Moreover, expression of only a small proportion of the cell identity genes was changed by the immunity response. Together, these data suggest that despite strong cell type specificity in its response, PTI does not affect cell identity networks.
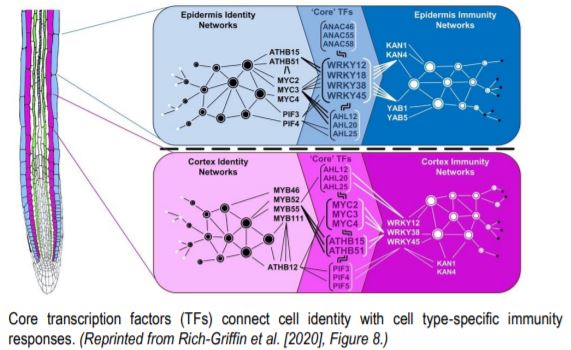
Next, Rich-Griffin et al. (2020) set out to identify which transcription factors connect cell identity and cell type-specific immunity networks. Given that control of gene expression often involves specific combinations of TF binding motifs (Lewis et al., 2015), the authors developed software called the Paired Motif Enrichment Tool (PMET) to identify such combinations. Using PMET, they identified two-motif combinations enriched in the promoters of cell identity genes and cell type-specific immunity genes. TFs acting in pairs with TFs from cell identity networks as well as cell type-specific immunity networks were assigned as core TFs, intersecting development and stress responses. In the epidermis, WRKY TFs (WRKY12/18/28/45) functioned as core TFs, acting in combination with ATHB (ARABIDOPSIS THALIANA HOMEOBOX), MYC and PIF (PHYTOCHROME INTERACTING FACTOR) TFs to regulate growth and development, while WRKYs combined with KAN (KANADI) and YAB (YABBY) TF networks to control epidermal immunity responses. In cortical cells, core TFs included MYC and ATHB TFs, whose binding sites were found in combination with cortex identity MYB binding elements, or with WRKY motifs in promoter regions of cortex-specific immunity genes.
The authors functionally validated a subset of identified motif pairs using a trans-activation YFP system. For example, native promoters with a WRKY-KAN motif co-activated YFP expression specifically in the epidermis, while replacing one of these motifs not only abolished basal YFP expression, but also prevented its induction by the PTI elicitor. Finally, the group designed a synthetic promoter containing the WRKY-KAN motif pair, which was sufficient to drive elicitor-induced YFP expression specifically in the epidermis. Narrowing down cell type-specific PTI to essential promoter elements and subsequent engineering of localized immune responses opens up exciting avenues for synthetic biology solutions to biotic stresses.
Together, Rich-Griffin et al. (2020) showed that immunity gene networks differ between root cell types and are interconnected with cell identity networks. Core TFs, acting as a “meeting point” for cell type‑specific immunity and cell identity networks, ensure that responses to pathogens are activated within the functional potential of individual root cell types. Sets of cell identity genes, whose cell type-specific expression is maintained under biotic (Rich-Griffin et al., 2020) or abiotic stresses (Iyer-Pascuzzi et al., 2011) preserve root cell identity under unfavorable conditions. It seems to be impossible to stress roots enough to cause their cells to lose their identity.
Dorota Kawa
Department of Plant Biology and Genome Center
University of California, Davis
ORCID: 0000-0002-4227-1621
REFERENCES
Bigeard J, Colcombet J, Hirt H (2015) Signaling mechanisms in pattern-triggered immunity (PTI). Mol Plant 8: 521-539
Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801-806
Iyer-Pascuzzi AS, Jackson T, Cui H, Petricka JJ, Busch W, Tsukagoshi H, Benfey PN (2011) Cell identity regulators link development and stress responses in the Arabidopsis root. Dev Cell 21: 770-782
Lewis LA, Polanski K, de Torres-Zabala M, Jayaraman S, Bowden L, Moore J, Penfold CA, Jenkins DJ, Hill C, Baxter L, Kulasekaran S, Truman W, Littlejohn G, Prusinska J, Mead A, Steinbrenner J, Hickman R, Rand D, Wild DL, Ott S, Buchanan-Wollaston V, Smirnoff N, Beynon J, Denby K, Grant M (2015) Transcriptional Dynamics Driving MAMP-Triggered Immunity and Pathogen Effector-Mediated Immunosuppression in Arabidopsis Leaves Following Infection with Pseudomonas syringae pv tomato DC3000. The Plant Cell 27: 3038-3064
Walker L, Boddington C, Jenkins D, Wang Y, Gronlund JT, Hulsmans J, Kumar S, Patel D, Moore JD, Carter A, Samavedam S, Bonomo G, Hersh DS, Coruzzi GM, Burroughs NJ, Gifford ML (2017) Changes in Gene Expression in Space and Time Orchestrate Environmentally Mediated Shaping of Root Architecture. Plant Cell 29: 2393-2412