Impact of Biolistic Transformation on the Genome
Liu et al. show that biolistic transformation can cause extensive damage and rearrangements, including deletions, duplications, chromosome fusions, and copy number variations. Plant Cell https://doi.org/10.1105/tpc.18.00613
By Jianing Liu and R. Kelly Dawe, Department of Genetics, University of Georgia.
Background: Agrobacterium-mediated gene transfer and biolistic transformation are the two most widely used methods for plant transformation. Agrobacterium transformation is the preferred method because it often delivers intact single-copy transgenes, whereas multicopy insertions and fragmented transgenes are prevalent outcomes with biolistic transformation. Despite its deficiencies, biolistic transformation has some notable advantages. It can be used in a wide range of species and can deliver any type of DNA, RNA or protein. Its flexibility has made it an attractive method for introducing Cas9 protein and repair templates in gene editing applications.
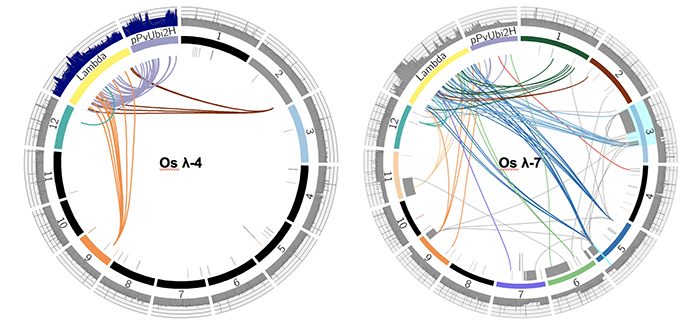
Question: We were initially interested in using biolistic transformation to transform long intact DNA molecules into rice and maize, and had chosen linear 48 kb phage lambda and smaller circular plasmids as test cases for this purpose. We wanted to use whole genome sequencing to see how transformation affected both the introduced DNA and the host genome.
Findings: We found that biolistic transformation can result in simple low copy insertions as well as large transgene arrays and severe genome rearrangements. Megabase-scale arrays of broken transgenes and genomic fragments were documented in multiple lines. There was evidence of severe genome damage, including large deletions, partial trisomy, and genome shattering events resembling chromothripsis and breakage-fusion bridge cycling. Other regions of the genome were broken and fragmented but seamlessly repaired. We conclude that transformation is the result of double strand breaks at the level of the transgene and the genome, and that known DNA repair pathways can explain the variety of outcomes observed.
Next steps: Our results highlight the fact that new methods of plant transformation are needed to make full use of emerging gene editing technologies. If biolistic methods are to be used, we recommend performing whole genome sequencing to identify lines without genomic rearrangements, or to outcross the transformants to normal breeding lines prior to detailed analysis.
Jianing Liu, Natalie J Nannas, Fang-Fang Fu, Jinghua Shi, Brooke Aspinwall, Wayne Allen Parrott, R. Kelly Dawe. (2019). Genome-scale Sequence Disruption Following Biolistic Transformation in Rice and Maize. Plant Cell DOI: https://doi.org/10.1105/tpc.18.00613
Key words/tags: genetics, genomics, plant transformation, technologies, gene editing.