Ethylene Represses Gene Transcription via Histone Deacetylases
Approximately half of all ethylene-responsive genes are downregulated in the presence in ethylene (Chang et al, 2013), but this repression has received relatively little attention compared to the ethylene-mediated activation of expression. The known positive regulators of ethylene signaling include ETHYLENE INSENSITIVE 2 (EIN2) and the transcription factor EIN3. Intriguingly, EIN3 binds to promoters of both ethylene-activated and -repressed genes. EIN2, by contrast, affects gene expression via its C-terminus, which is cleaved in the presence of ethylene and translocated to the nucleus, where it was recently shown to alter HISTONE 3 acetylation levels in a process that involves EIN2 NUCLEAR ASSOCIATED PROTEIN 1 (ENAP1; Zhang et al., 2016; Zhang et al., 2017). Now, Zhang et al. (2018) report that two histone deacetylases function with ENAP1 to repress gene expression in the presence of ethylene.
The authors aimed determine which histone deacetylases (HDACs) might be involved in the EIN2- and ENAP1-mediated regulation of histone acetylation. They tested the 18 known HDACs of Arabidopsis thaliana for interaction with ENAP1 or the C-terminal region of EIN2 and found that SIRTUIN 1 (SRT1) and SRT2 interacted with ENAP1 (none interacted with EIN2). These interactions with ENAP1 were enhanced upon ethylene treatment. When the authors examined single mutants of srt1 and srt2, they found evidence of a compensatory mechanism wherein the remaining gene was upregulated, and neither mutant showed clear ethylene-related phenotypes. The srt1 srt2 double mutant, however, showed decreased responsiveness to ethylene. Conversely, overexpression of SRT2 led to a degree of ethylene hypersensitivity. Thus, Zhang et al. demonstrated that SRT1 and SRT2 are indeed involved in plant responses to ethylene, as their interaction with ENAP1 suggested.
As SRT1 and SRT2 were predicted to be histone deacetylases, and therefore presumably function by influencing gene expression, the authors used transcriptomic analyses to explore their roles in the regulation of gene expression. Using the srt1 srt2 double mutant, Zhang et al. found that 35% of the ethylene-responsive genes from wild type were SRT1/SRT2-regulated. Notably, among them, 50% of the genes that were repressed by ethylene in the wild type were no longer repressed by ethylene treatment in the srt1 srt2 double mutant, indicating that SRT1 and/or SRT2 mediate the repression of these genes. This conclusion was supported by the finding that the same genes were downregulated in an SRT2-overexpression line, even without ethylene treatment.
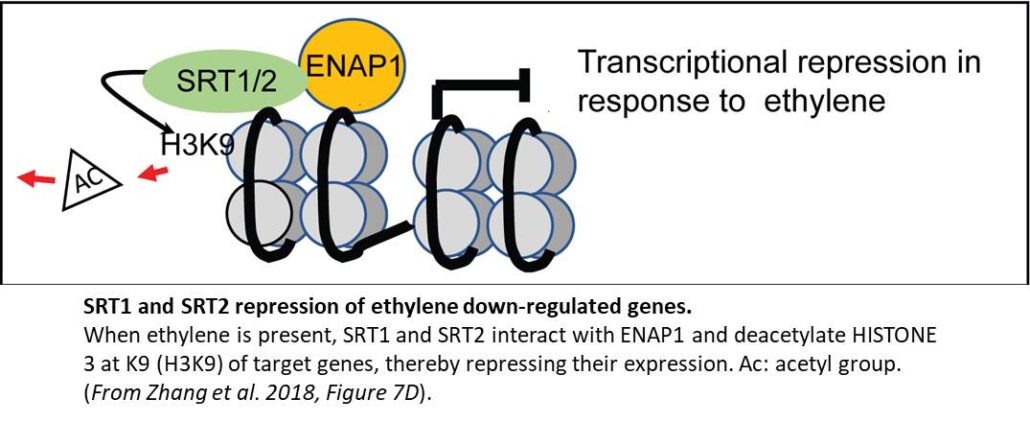
The authors confirmed that SRT2 has histone deacetylase activity and showed that SRT1 and SRT2 influence the acetylation of HISTONE 3 on K9 in planta. Furthermore, chromatin immunoprecipitation assays showed both ENAP1 and SRT2 bound to promoters of ethylene-repressed genes, with SRT2 binding increased in the presence of ethylene (see Figure). In addition, the srt1 srt2 double mutant background rescued the ethylene hypersensitivity caused by overexpression of ENAP1—with expression of ethylene-repressed genes specifically affected—providing evidence that SRT1 and SRT2 function in the same pathway as ENAP1.
Finally, the authors explored the relationship between SRT1/SRT2 function and EIN3-mediated regulation of ethylene-responsive genes, finding that the srt1 srt2 double mutant background rescued the ethylene hypersensitivity caused by overexpression of EIN3. Notably, this rescue was specifically associated with altered expression of ethylene-repressed genes.
These findings from Chang et al. highlight both the roles of SRT1 and SRT2 in ethylene repression of gene expression and the importance of this repression to ethylene responses within the plant. This work thus provides good evidence linking the modulation of chromatin architecture to hormone responses.
References
Chang, K.N., Zhong, S., Weirauch, M.T., Hon, G., Pelizzola, M., Li, H., Huang, S.S., Schmitz, R.J., Urich, M.A., Kuo, D., Nery, J.R., Qiao, H., Yang, A., Jamali, A., Chen, H., Ideker, T., Ren, B., Bar-Joseph, Z., Hughes, T.R., and Ecker, J.R. (2013). Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. Elife 2, e00675.
Zhang, F., Qi, B., Wang, L.K., Zhao, B., Rode, S., Riggan, N.D., Ecker, J.R., and Qiao, H. (2016). EIN2-dependent regulation of acetylation of histone H3K14 and non-canonical histone H3K23 in ethylene signalling. Nat Commun 7, 13018.
Zhang, F., Wang, L., Ko, E.E., Shao, K., and Qiao, H. (2018). Histone Deacetylases SRT1 and SRT2 Interact with ENAP1 To Mediate Ethylene-Induced Transcriptional Repression. Plant Cell. doi: 10.1105/tpc.17.00671.
Zhang, F., Wang, L., Qi, B., Zhao, B., Ko, E.E., Riggan, N.D., Chin, K., and Qiao, H. (2017). EIN2 mediates direct regulation of histone acetylation in the ethylene response. Proc Natl Acad Sci U S A 114, 10274-10279.