Development of genome-wide SNP markers for barley via reference- based RNA-Seq Analysis (Front Plant Sci)
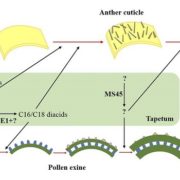
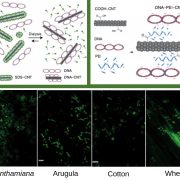
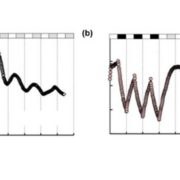
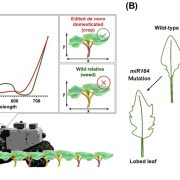
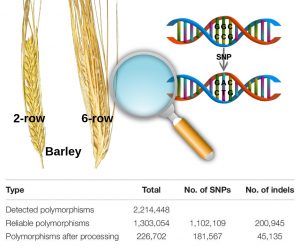 The availability of marker-assisted systems is key for crop improvement, allowing trait selection by identifying consensus polymorphisms. However, for discriminating between related strains, these DNA markers are limited, time-consuming and expensive. Here, Tanaka et al. developed a RNA-Seq-based genotyping pipeline for genome-wide polymorphism detection for barley. The authors compared the variations among 108 Japanese strains of two and six-row barley, by mapping onto the reference public genome sequence of cultivar Morex (Hordeum vulgare) to test efficiency. Using only the transcript region and their ∼3-Kbp flanking regions, instead of the whole genome, reduces the mapping time by half and the genotyping time by two-thirds. They validated this method by amplicon sequencing (AmpliSeq) and found that AmpliSeq detects a wider variety of polymorphisms than RNA-Seq, yet it requires prior identification of the polymorphisms; thus an optimal approach is the combination of both techniques. This study reveals the efficiency reached by a DNA marker based on RNA-Seq able to compare related strains. As expected, the number of polymorphisms was lower in same than between different row type strains. (Summary by Ana Valladares). Front Plant Sci. 10.3389/fpls.2019.00403
The availability of marker-assisted systems is key for crop improvement, allowing trait selection by identifying consensus polymorphisms. However, for discriminating between related strains, these DNA markers are limited, time-consuming and expensive. Here, Tanaka et al. developed a RNA-Seq-based genotyping pipeline for genome-wide polymorphism detection for barley. The authors compared the variations among 108 Japanese strains of two and six-row barley, by mapping onto the reference public genome sequence of cultivar Morex (Hordeum vulgare) to test efficiency. Using only the transcript region and their ∼3-Kbp flanking regions, instead of the whole genome, reduces the mapping time by half and the genotyping time by two-thirds. They validated this method by amplicon sequencing (AmpliSeq) and found that AmpliSeq detects a wider variety of polymorphisms than RNA-Seq, yet it requires prior identification of the polymorphisms; thus an optimal approach is the combination of both techniques. This study reveals the efficiency reached by a DNA marker based on RNA-Seq able to compare related strains. As expected, the number of polymorphisms was lower in same than between different row type strains. (Summary by Ana Valladares). Front Plant Sci. 10.3389/fpls.2019.00403