CrY2H-seq: a massively multiplexed assay for deep-coverage interactome mapping ($)
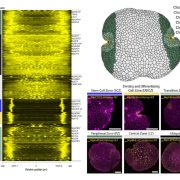
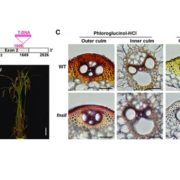
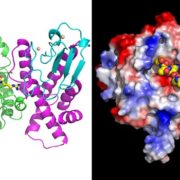
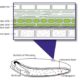
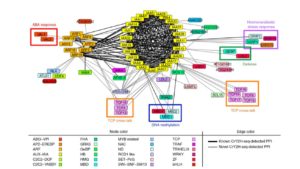 Protein-protein interactions are crucial to our understanding of biology but can be hard to detect. Trigg et al. developed a sophisticated yeast two-hybrid assay augmented with Cre recombinase (CrY2H-seq) to identify the Arabidopsis transcription factor protein-protein interactome. In this method, plasmid libraries carrying transcription factor open-reading frames (ORFs) fused to GAL4 activation domains or DNA-binding domains, flanked by lox sequences (targets of Cre recombinase) are introduced into yeast cells with a genome-integrated Cre recombinase ORF under the control of the GAL7 promoter. Interaction in the yeast cells between the two proteins leads to expression of Cre recombinase, which targets the lox sequences and joins the two plasmids together. Plasmid recovery and sequencing reveals pairs of interacting proteins. Starting with nearly 2000 Arabidopsis transcription factors, the authors identified nearly 9000 novel and known interactions and independently validated hundreds of them. Biological functions implicated by the uncovered interactions include gynoecium development, phosphate sensing and circadian coordination. With CrY2H-seq facilitating massively-multiplexed interactome screening, a more complete arabidopsis interactome as well as potential interactomes for other plants are on the horizon. The searchable dataset ‘Arabidopsis thaliana transcription factor interaction network, version 1’ (AtTFIN-1) is available online (http://signal.salk.edu/interactome/AtTFIN-1.html). Nature Methods 10.1038/nmeth.4343
Protein-protein interactions are crucial to our understanding of biology but can be hard to detect. Trigg et al. developed a sophisticated yeast two-hybrid assay augmented with Cre recombinase (CrY2H-seq) to identify the Arabidopsis transcription factor protein-protein interactome. In this method, plasmid libraries carrying transcription factor open-reading frames (ORFs) fused to GAL4 activation domains or DNA-binding domains, flanked by lox sequences (targets of Cre recombinase) are introduced into yeast cells with a genome-integrated Cre recombinase ORF under the control of the GAL7 promoter. Interaction in the yeast cells between the two proteins leads to expression of Cre recombinase, which targets the lox sequences and joins the two plasmids together. Plasmid recovery and sequencing reveals pairs of interacting proteins. Starting with nearly 2000 Arabidopsis transcription factors, the authors identified nearly 9000 novel and known interactions and independently validated hundreds of them. Biological functions implicated by the uncovered interactions include gynoecium development, phosphate sensing and circadian coordination. With CrY2H-seq facilitating massively-multiplexed interactome screening, a more complete arabidopsis interactome as well as potential interactomes for other plants are on the horizon. The searchable dataset ‘Arabidopsis thaliana transcription factor interaction network, version 1’ (AtTFIN-1) is available online (http://signal.salk.edu/interactome/AtTFIN-1.html). Nature Methods 10.1038/nmeth.4343