Chromatin signature and transcription factor binding provide a predictive basis for understanding plant gene expression (Plant Cell Physiol)
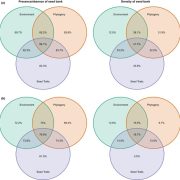
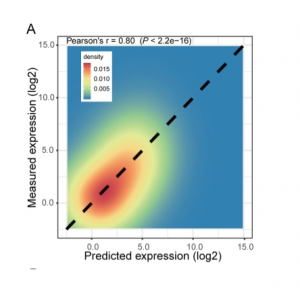 Machine learning is a booming research field, also in Plant Science. Here, Wu et al. use chromatin modifications and transcription factors to predict transcription levels in Arabidopsis and rice. This is not only important for prediction but also to understand the mechanisms underlying epigenetic regulation. For chromatin modifications, they studied the combinatorial effect of histone modification and DNaseI hypersensitivity, producing good predictive power. According to their model, H3K4me3, H3K36me3, H3K9ac and H3K27ac are the best predictors of expression. Moreover, trained models could be applied to different tissues or species evidencing conserved mechanisms. In the case of transcription-factor based models, the authors describe that binding in the promoter region only has some predictive power in a few cases, showing that there is still work to do on this subject. Overall, this paper is important for the establishment of methods to deal with this kind of predictive problems. The authors also provide a description of plant gene expression regulation features that evidence some known and other novel correlations. (Summary by Facundo Romani) Plant Cell Physiol. 10.1093/pcp/pcz051/5436135
Machine learning is a booming research field, also in Plant Science. Here, Wu et al. use chromatin modifications and transcription factors to predict transcription levels in Arabidopsis and rice. This is not only important for prediction but also to understand the mechanisms underlying epigenetic regulation. For chromatin modifications, they studied the combinatorial effect of histone modification and DNaseI hypersensitivity, producing good predictive power. According to their model, H3K4me3, H3K36me3, H3K9ac and H3K27ac are the best predictors of expression. Moreover, trained models could be applied to different tissues or species evidencing conserved mechanisms. In the case of transcription-factor based models, the authors describe that binding in the promoter region only has some predictive power in a few cases, showing that there is still work to do on this subject. Overall, this paper is important for the establishment of methods to deal with this kind of predictive problems. The authors also provide a description of plant gene expression regulation features that evidence some known and other novel correlations. (Summary by Facundo Romani) Plant Cell Physiol. 10.1093/pcp/pcz051/5436135