Breakthrough in the identification of photosynthesis genes in green algae
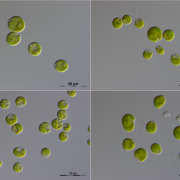
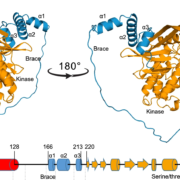
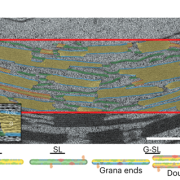
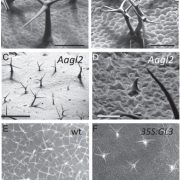
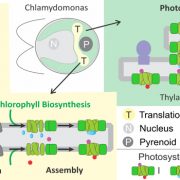
 Imagine the everyday delights of foods like bread, ramen, or sushi. Central to these culinary staples is starch, a product of the photosynthesis process in plants. Despite its critical role, people still do not fully understand the regulation and biogenesis of the photosynthesis machinery. In this study, Kafri et al. identified 70 previously unknown photosynthesis genes in the green algae species Chlamydomonas reinhardtii, offering deeper understanding of this process. Both wild-type and photosynthesis-deficient Chlamydomonas can grow in dark environments when supplied with acetate as a carbon source, but in light conditions without acetate, photosynthesis-deficient mutants display notable growth defects. Leveraging this characteristic, the authors conducted a high-throughput automated spot test on agar to verify a set of 1,781 previously identified photosynthesis-deficient Chlamydomonas mutants. Subsequently, the authors designed a pooled backcrossing strategy to map the causal mutations and narrowed the pool down to 115 high fidelity genes, including 70 previously uncharacterized genes. To validate their findings, the team performed genetic complementation for some of the hits and examined their protein localization. Further, a large-scale proteomic profiling of these mutants informed the impact of the loss of each gene on the proteome, with some causing strong depletion of the photosynthetic complexes, indicating their regulatory roles. Notably, two master regulators of photosynthesis, MTF1 and PMR1, were identified. This study marks a significant leap forward for the understanding of the photosynthetic machinery in Chlamydomonas, and provides an important resource to pave the way for further explorations of photosynthesis. (Summary by Xiaohui Li @Xiao_hui_Li) Cell 10.1016/j.cell.2023.11.007
Imagine the everyday delights of foods like bread, ramen, or sushi. Central to these culinary staples is starch, a product of the photosynthesis process in plants. Despite its critical role, people still do not fully understand the regulation and biogenesis of the photosynthesis machinery. In this study, Kafri et al. identified 70 previously unknown photosynthesis genes in the green algae species Chlamydomonas reinhardtii, offering deeper understanding of this process. Both wild-type and photosynthesis-deficient Chlamydomonas can grow in dark environments when supplied with acetate as a carbon source, but in light conditions without acetate, photosynthesis-deficient mutants display notable growth defects. Leveraging this characteristic, the authors conducted a high-throughput automated spot test on agar to verify a set of 1,781 previously identified photosynthesis-deficient Chlamydomonas mutants. Subsequently, the authors designed a pooled backcrossing strategy to map the causal mutations and narrowed the pool down to 115 high fidelity genes, including 70 previously uncharacterized genes. To validate their findings, the team performed genetic complementation for some of the hits and examined their protein localization. Further, a large-scale proteomic profiling of these mutants informed the impact of the loss of each gene on the proteome, with some causing strong depletion of the photosynthetic complexes, indicating their regulatory roles. Notably, two master regulators of photosynthesis, MTF1 and PMR1, were identified. This study marks a significant leap forward for the understanding of the photosynthetic machinery in Chlamydomonas, and provides an important resource to pave the way for further explorations of photosynthesis. (Summary by Xiaohui Li @Xiao_hui_Li) Cell 10.1016/j.cell.2023.11.007