Base-editing in RNA and DNA
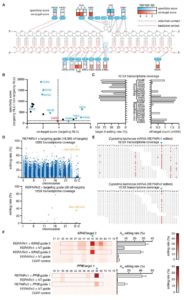 The ability to engineer precise changes in nucleic acid sequences has advanced rapidly over the last few years. Since the development of genome editing technologies such as CRISPR-Cas9, a modified version known as base editing has sought to reliably convert individual nucleotides. All known base editing techniques harness cytidine deaminases fused to a catalytically inactive Cas9 to permit C•G to T•A conversions. Gaudelli et al. (Nature 10.1038/nature24644) created a modified version of an E. coli adenosine deaminase through several rounds of bacterial evolution that was able to mediate specific A•T to G•C conversions at high efficiency while minimizing off-target effects. As DNA base editors can target both the sense and anti-sense strands, it is now possible to mediate all four base transitions in genomic DNA.
The ability to engineer precise changes in nucleic acid sequences has advanced rapidly over the last few years. Since the development of genome editing technologies such as CRISPR-Cas9, a modified version known as base editing has sought to reliably convert individual nucleotides. All known base editing techniques harness cytidine deaminases fused to a catalytically inactive Cas9 to permit C•G to T•A conversions. Gaudelli et al. (Nature 10.1038/nature24644) created a modified version of an E. coli adenosine deaminase through several rounds of bacterial evolution that was able to mediate specific A•T to G•C conversions at high efficiency while minimizing off-target effects. As DNA base editors can target both the sense and anti-sense strands, it is now possible to mediate all four base transitions in genomic DNA.
Cox et al. (Science 10.1126/science.aaq0180) used a similar approach to repurpose the RNA-binding Cas13 protein to perform base editing within RNA transcripts. They developed a version of Cas13 without nuclease activity and fused it to an adenosine deaminase acting on RNA (ADAR) family protein. This fusion was capable of directing adenosine to inosine (read as guanosine) replacements within a target site. The ADAR protein was subject to bacterial evolution to minimize non-specific edits around the target site. While currently lacking the range of DNA base editors, the non-permanency of RNA editing is advantageous for some applications.
These tools are likely to help shape agronomic research in the near future. (Summaries by Mike Page).